Jason Alan Fries
MedHELM: Holistic Evaluation of Large Language Models for Medical Tasks
May 26, 2025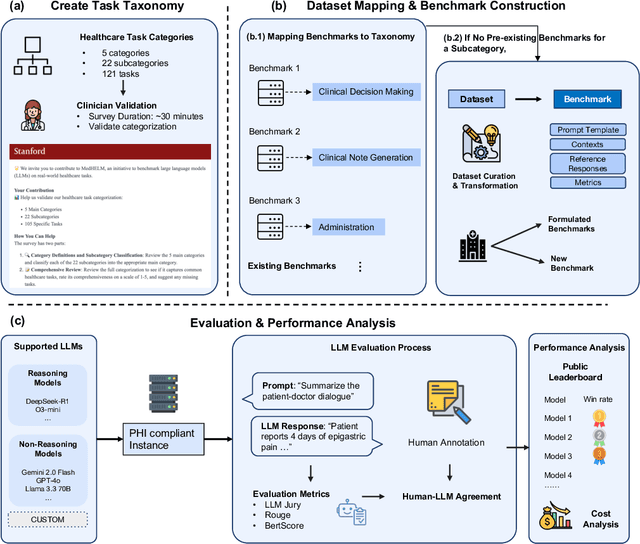
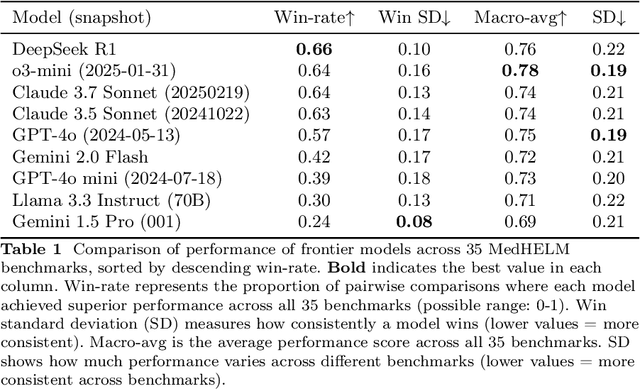
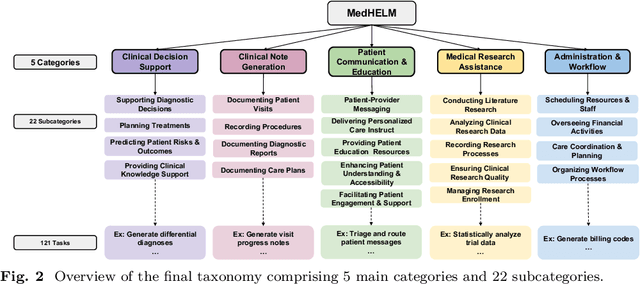
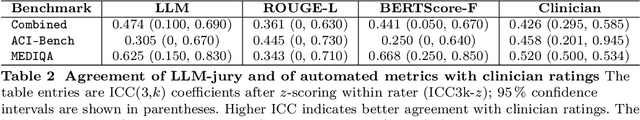
Abstract:While large language models (LLMs) achieve near-perfect scores on medical licensing exams, these evaluations inadequately reflect the complexity and diversity of real-world clinical practice. We introduce MedHELM, an extensible evaluation framework for assessing LLM performance for medical tasks with three key contributions. First, a clinician-validated taxonomy spanning 5 categories, 22 subcategories, and 121 tasks developed with 29 clinicians. Second, a comprehensive benchmark suite comprising 35 benchmarks (17 existing, 18 newly formulated) providing complete coverage of all categories and subcategories in the taxonomy. Third, a systematic comparison of LLMs with improved evaluation methods (using an LLM-jury) and a cost-performance analysis. Evaluation of 9 frontier LLMs, using the 35 benchmarks, revealed significant performance variation. Advanced reasoning models (DeepSeek R1: 66% win-rate; o3-mini: 64% win-rate) demonstrated superior performance, though Claude 3.5 Sonnet achieved comparable results at 40% lower estimated computational cost. On a normalized accuracy scale (0-1), most models performed strongly in Clinical Note Generation (0.73-0.85) and Patient Communication & Education (0.78-0.83), moderately in Medical Research Assistance (0.65-0.75), and generally lower in Clinical Decision Support (0.56-0.72) and Administration & Workflow (0.53-0.63). Our LLM-jury evaluation method achieved good agreement with clinician ratings (ICC = 0.47), surpassing both average clinician-clinician agreement (ICC = 0.43) and automated baselines including ROUGE-L (0.36) and BERTScore-F1 (0.44). Claude 3.5 Sonnet achieved comparable performance to top models at lower estimated cost. These findings highlight the importance of real-world, task-specific evaluation for medical use of LLMs and provides an open source framework to enable this.
TIMER: Temporal Instruction Modeling and Evaluation for Longitudinal Clinical Records
Mar 06, 2025Abstract:Large language models (LLMs) have emerged as promising tools for assisting in medical tasks, yet processing Electronic Health Records (EHRs) presents unique challenges due to their longitudinal nature. While LLMs' capabilities to perform medical tasks continue to improve, their ability to reason over temporal dependencies across multiple patient visits and time frames remains unexplored. We introduce TIMER (Temporal Instruction Modeling and Evaluation for Longitudinal Clinical Records), a framework that incorporate instruction-response pairs grounding to different parts of a patient's record as a critical dimension in both instruction evaluation and tuning for longitudinal clinical records. We develop TIMER-Bench, the first time-aware benchmark that evaluates temporal reasoning capabilities over longitudinal EHRs, as well as TIMER-Instruct, an instruction-tuning methodology for LLMs to learn reasoning over time. We demonstrate that models fine-tuned with TIMER-Instruct improve performance by 7.3% on human-generated benchmarks and 9.2% on TIMER-Bench, indicating that temporal instruction-tuning improves model performance for reasoning over EHR.
Assessing the Limitations of Large Language Models in Clinical Fact Decomposition
Dec 17, 2024Abstract:Verifying factual claims is critical for using large language models (LLMs) in healthcare. Recent work has proposed fact decomposition, which uses LLMs to rewrite source text into concise sentences conveying a single piece of information, as an approach for fine-grained fact verification. Clinical documentation poses unique challenges for fact decomposition due to dense terminology and diverse note types. To explore these challenges, we present FactEHR, a dataset consisting of full document fact decompositions for 2,168 clinical notes spanning four types from three hospital systems. Our evaluation, including review by clinicians, highlights significant variability in the quality of fact decomposition for four commonly used LLMs, with some LLMs generating 2.6x more facts per sentence than others. The results underscore the need for better LLM capabilities to support factual verification in clinical text. To facilitate future research in this direction, we plan to release our code at \url{https://github.com/som-shahlab/factehr}.
Context Clues: Evaluating Long Context Models for Clinical Prediction Tasks on EHRs
Dec 09, 2024



Abstract:Foundation Models (FMs) trained on Electronic Health Records (EHRs) have achieved state-of-the-art results on numerous clinical prediction tasks. However, most existing EHR FMs have context windows of <1k tokens. This prevents them from modeling full patient EHRs which can exceed 10k's of events. Recent advancements in subquadratic long-context architectures (e.g., Mamba) offer a promising solution. However, their application to EHR data has not been well-studied. We address this gap by presenting the first systematic evaluation of the effect of context length on modeling EHR data. We find that longer context models improve predictive performance -- our Mamba-based model surpasses the prior state-of-the-art on 9/14 tasks on the EHRSHOT prediction benchmark. For clinical applications, however, model performance alone is insufficient -- robustness to the unique properties of EHR is crucial. Thus, we also evaluate models across three previously underexplored properties of EHR data: (1) the prevalence of "copy-forwarded" diagnoses which creates artificial repetition of tokens within EHR sequences; (2) the irregular time intervals between EHR events which can lead to a wide range of timespans within a context window; and (3) the natural increase in disease complexity over time which makes later tokens in the EHR harder to predict than earlier ones. Stratifying our EHRSHOT results, we find that higher levels of each property correlate negatively with model performance, but that longer context models are more robust to more extreme levels of these properties. Our work highlights the potential for using long-context architectures to model EHR data, and offers a case study for identifying new challenges in modeling sequential data motivated by domains outside of natural language. We release our models and code at: https://github.com/som-shahlab/long_context_clues
Time-to-Event Pretraining for 3D Medical Imaging
Nov 14, 2024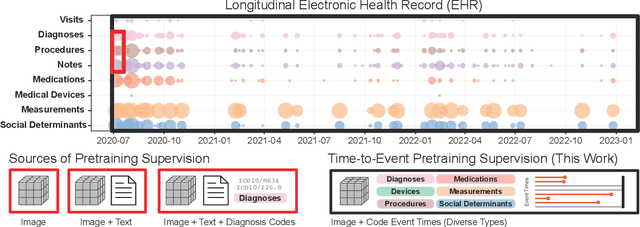
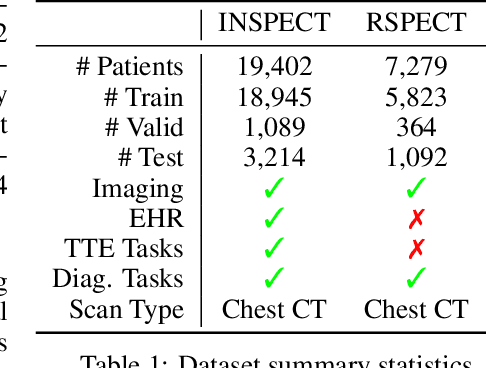
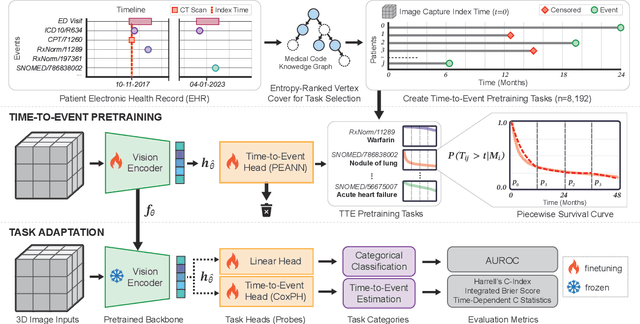
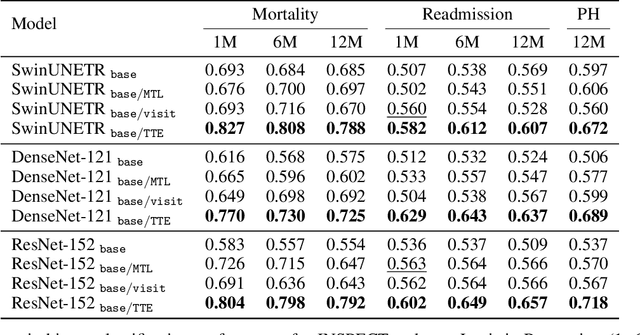
Abstract:With the rise of medical foundation models and the growing availability of imaging data, scalable pretraining techniques offer a promising way to identify imaging biomarkers predictive of future disease risk. While current self-supervised methods for 3D medical imaging models capture local structural features like organ morphology, they fail to link pixel biomarkers with long-term health outcomes due to a missing context problem. Current approaches lack the temporal context necessary to identify biomarkers correlated with disease progression, as they rely on supervision derived only from images and concurrent text descriptions. To address this, we introduce time-to-event pretraining, a pretraining framework for 3D medical imaging models that leverages large-scale temporal supervision from paired, longitudinal electronic health records (EHRs). Using a dataset of 18,945 CT scans (4.2 million 2D images) and time-to-event distributions across thousands of EHR-derived tasks, our method improves outcome prediction, achieving an average AUROC increase of 23.7% and a 29.4% gain in Harrell's C-index across 8 benchmark tasks. Importantly, these gains are achieved without sacrificing diagnostic classification performance. This study lays the foundation for integrating longitudinal EHR and 3D imaging data to advance clinical risk prediction.
meds_reader: A fast and efficient EHR processing library
Sep 12, 2024



Abstract:The growing demand for machine learning in healthcare requires processing increasingly large electronic health record (EHR) datasets, but existing pipelines are not computationally efficient or scalable. In this paper, we introduce meds_reader, an optimized Python package for efficient EHR data processing that is designed to take advantage of many intrinsic properties of EHR data for improved speed. We then demonstrate the benefits of meds_reader by reimplementing key components of two major EHR processing pipelines, achieving 10-100x improvements in memory, speed, and disk usage. The code for meds_reader can be found at https://github.com/som-shahlab/meds_reader.
BLOOM: A 176B-Parameter Open-Access Multilingual Language Model
Nov 09, 2022Abstract:Large language models (LLMs) have been shown to be able to perform new tasks based on a few demonstrations or natural language instructions. While these capabilities have led to widespread adoption, most LLMs are developed by resource-rich organizations and are frequently kept from the public. As a step towards democratizing this powerful technology, we present BLOOM, a 176B-parameter open-access language model designed and built thanks to a collaboration of hundreds of researchers. BLOOM is a decoder-only Transformer language model that was trained on the ROOTS corpus, a dataset comprising hundreds of sources in 46 natural and 13 programming languages (59 in total). We find that BLOOM achieves competitive performance on a wide variety of benchmarks, with stronger results after undergoing multitask prompted finetuning. To facilitate future research and applications using LLMs, we publicly release our models and code under the Responsible AI License.
BigBIO: A Framework for Data-Centric Biomedical Natural Language Processing
Jun 30, 2022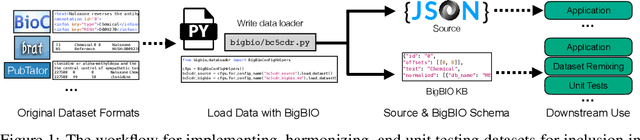
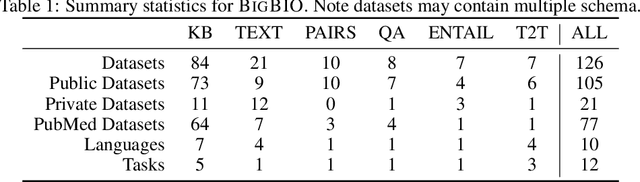
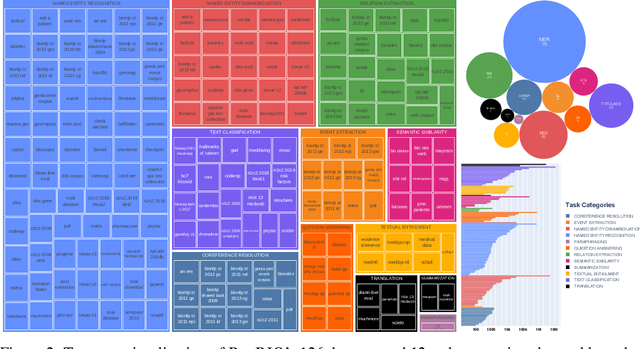
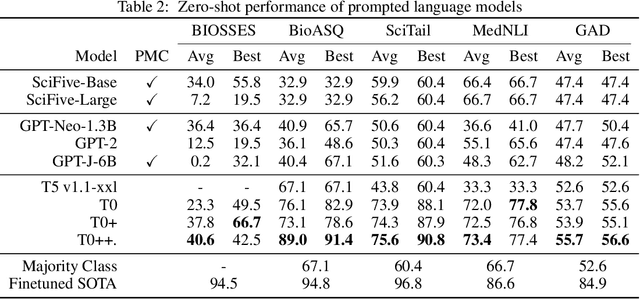
Abstract:Training and evaluating language models increasingly requires the construction of meta-datasets --diverse collections of curated data with clear provenance. Natural language prompting has recently lead to improved zero-shot generalization by transforming existing, supervised datasets into a diversity of novel pretraining tasks, highlighting the benefits of meta-dataset curation. While successful in general-domain text, translating these data-centric approaches to biomedical language modeling remains challenging, as labeled biomedical datasets are significantly underrepresented in popular data hubs. To address this challenge, we introduce BigBIO a community library of 126+ biomedical NLP datasets, currently covering 12 task categories and 10+ languages. BigBIO facilitates reproducible meta-dataset curation via programmatic access to datasets and their metadata, and is compatible with current platforms for prompt engineering and end-to-end few/zero shot language model evaluation. We discuss our process for task schema harmonization, data auditing, contribution guidelines, and outline two illustrative use cases: zero-shot evaluation of biomedical prompts and large-scale, multi-task learning. BigBIO is an ongoing community effort and is available at https://github.com/bigscience-workshop/biomedical
PromptSource: An Integrated Development Environment and Repository for Natural Language Prompts
Feb 02, 2022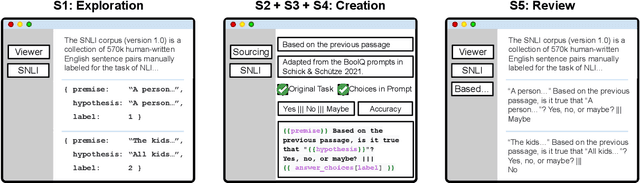
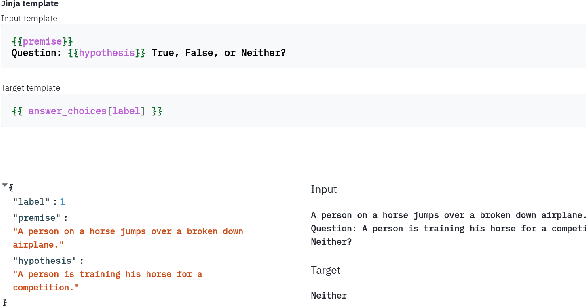

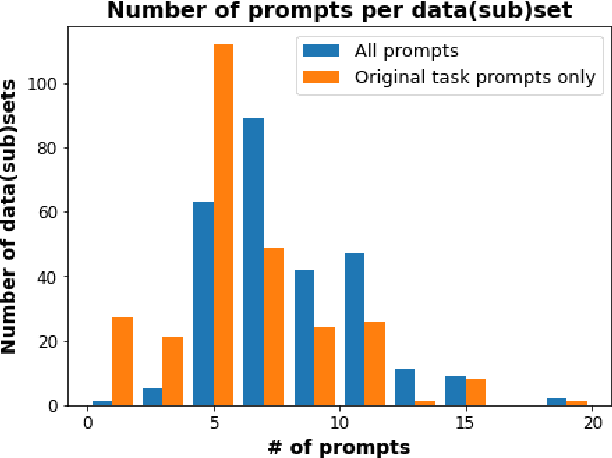
Abstract:PromptSource is a system for creating, sharing, and using natural language prompts. Prompts are functions that map an example from a dataset to a natural language input and target output. Using prompts to train and query language models is an emerging area in NLP that requires new tools that let users develop and refine these prompts collaboratively. PromptSource addresses the emergent challenges in this new setting with (1) a templating language for defining data-linked prompts, (2) an interface that lets users quickly iterate on prompt development by observing outputs of their prompts on many examples, and (3) a community-driven set of guidelines for contributing new prompts to a common pool. Over 2,000 prompts for roughly 170 datasets are already available in PromptSource. PromptSource is available at https://github.com/bigscience-workshop/promptsource.
RadFusion: Benchmarking Performance and Fairness for Multimodal Pulmonary Embolism Detection from CT and EHR
Nov 27, 2021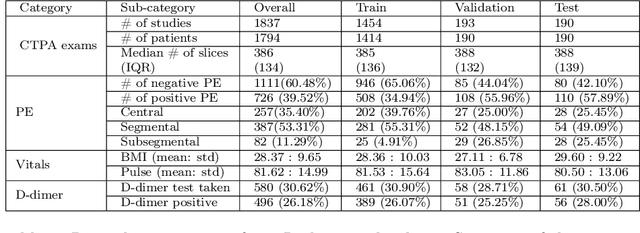
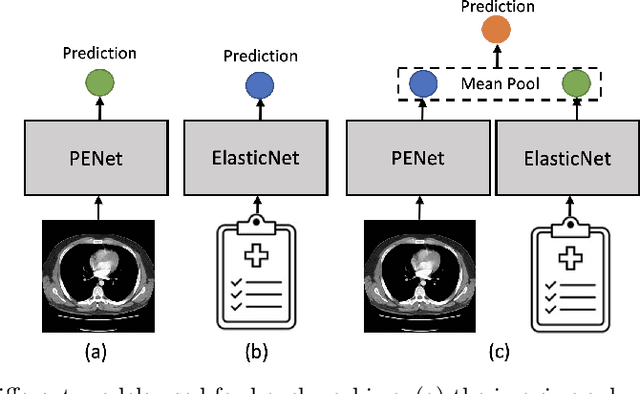
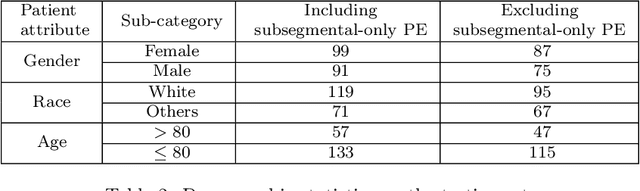
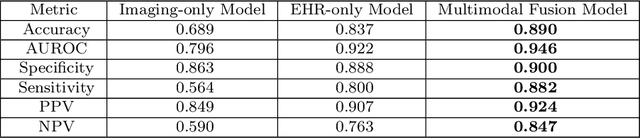
Abstract:Despite the routine use of electronic health record (EHR) data by radiologists to contextualize clinical history and inform image interpretation, the majority of deep learning architectures for medical imaging are unimodal, i.e., they only learn features from pixel-level information. Recent research revealing how race can be recovered from pixel data alone highlights the potential for serious biases in models which fail to account for demographics and other key patient attributes. Yet the lack of imaging datasets which capture clinical context, inclusive of demographics and longitudinal medical history, has left multimodal medical imaging underexplored. To better assess these challenges, we present RadFusion, a multimodal, benchmark dataset of 1794 patients with corresponding EHR data and high-resolution computed tomography (CT) scans labeled for pulmonary embolism. We evaluate several representative multimodal fusion models and benchmark their fairness properties across protected subgroups, e.g., gender, race/ethnicity, age. Our results suggest that integrating imaging and EHR data can improve classification performance and robustness without introducing large disparities in the true positive rate between population groups.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge