Ethan Steinberg
Context Clues: Evaluating Long Context Models for Clinical Prediction Tasks on EHRs
Dec 09, 2024



Abstract:Foundation Models (FMs) trained on Electronic Health Records (EHRs) have achieved state-of-the-art results on numerous clinical prediction tasks. However, most existing EHR FMs have context windows of <1k tokens. This prevents them from modeling full patient EHRs which can exceed 10k's of events. Recent advancements in subquadratic long-context architectures (e.g., Mamba) offer a promising solution. However, their application to EHR data has not been well-studied. We address this gap by presenting the first systematic evaluation of the effect of context length on modeling EHR data. We find that longer context models improve predictive performance -- our Mamba-based model surpasses the prior state-of-the-art on 9/14 tasks on the EHRSHOT prediction benchmark. For clinical applications, however, model performance alone is insufficient -- robustness to the unique properties of EHR is crucial. Thus, we also evaluate models across three previously underexplored properties of EHR data: (1) the prevalence of "copy-forwarded" diagnoses which creates artificial repetition of tokens within EHR sequences; (2) the irregular time intervals between EHR events which can lead to a wide range of timespans within a context window; and (3) the natural increase in disease complexity over time which makes later tokens in the EHR harder to predict than earlier ones. Stratifying our EHRSHOT results, we find that higher levels of each property correlate negatively with model performance, but that longer context models are more robust to more extreme levels of these properties. Our work highlights the potential for using long-context architectures to model EHR data, and offers a case study for identifying new challenges in modeling sequential data motivated by domains outside of natural language. We release our models and code at: https://github.com/som-shahlab/long_context_clues
Time-to-Event Pretraining for 3D Medical Imaging
Nov 14, 2024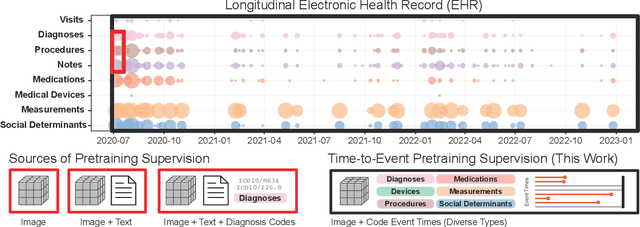
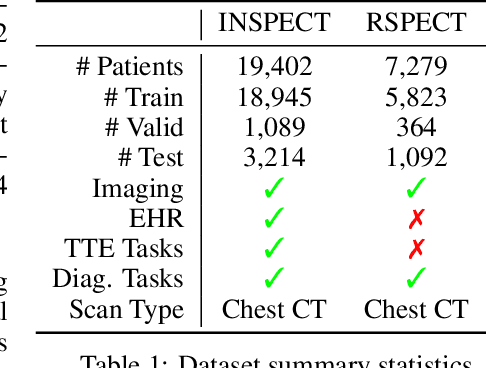
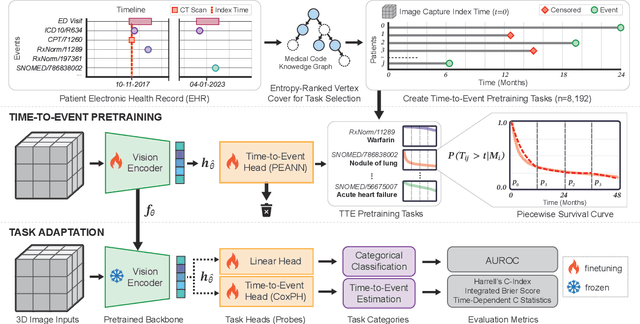
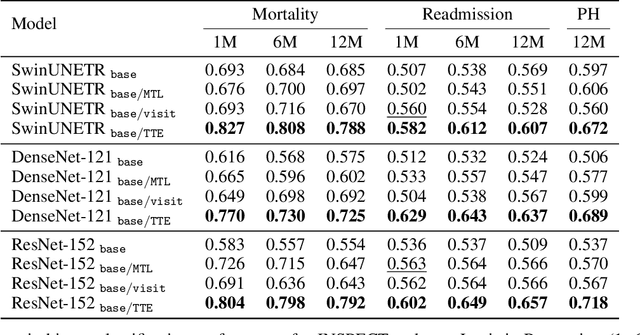
Abstract:With the rise of medical foundation models and the growing availability of imaging data, scalable pretraining techniques offer a promising way to identify imaging biomarkers predictive of future disease risk. While current self-supervised methods for 3D medical imaging models capture local structural features like organ morphology, they fail to link pixel biomarkers with long-term health outcomes due to a missing context problem. Current approaches lack the temporal context necessary to identify biomarkers correlated with disease progression, as they rely on supervision derived only from images and concurrent text descriptions. To address this, we introduce time-to-event pretraining, a pretraining framework for 3D medical imaging models that leverages large-scale temporal supervision from paired, longitudinal electronic health records (EHRs). Using a dataset of 18,945 CT scans (4.2 million 2D images) and time-to-event distributions across thousands of EHR-derived tasks, our method improves outcome prediction, achieving an average AUROC increase of 23.7% and a 29.4% gain in Harrell's C-index across 8 benchmark tasks. Importantly, these gains are achieved without sacrificing diagnostic classification performance. This study lays the foundation for integrating longitudinal EHR and 3D imaging data to advance clinical risk prediction.
meds_reader: A fast and efficient EHR processing library
Sep 12, 2024



Abstract:The growing demand for machine learning in healthcare requires processing increasingly large electronic health record (EHR) datasets, but existing pipelines are not computationally efficient or scalable. In this paper, we introduce meds_reader, an optimized Python package for efficient EHR data processing that is designed to take advantage of many intrinsic properties of EHR data for improved speed. We then demonstrate the benefits of meds_reader by reimplementing key components of two major EHR processing pipelines, achieving 10-100x improvements in memory, speed, and disk usage. The code for meds_reader can be found at https://github.com/som-shahlab/meds_reader.
A Multi-Center Study on the Adaptability of a Shared Foundation Model for Electronic Health Records
Nov 20, 2023Abstract:Foundation models hold promise for transforming AI in healthcare by providing modular components that are easily adaptable to downstream healthcare tasks, making AI development more scalable and cost-effective. Structured EHR foundation models, trained on coded medical records from millions of patients, demonstrated benefits including increased performance with fewer training labels, and improved robustness to distribution shifts. However, questions remain on the feasibility of sharing these models across different hospitals and their performance for local task adaptation. This multi-center study examined the adaptability of a recently released structured EHR foundation model ($FM_{SM}$), trained on longitudinal medical record data from 2.57M Stanford Medicine patients. Experiments were conducted using EHR data at The Hospital for Sick Children and MIMIC-IV. We assessed both adaptability via continued pretraining on local data, and task adaptability compared to baselines of training models from scratch at each site, including a local foundation model. We evaluated the performance of these models on 8 clinical prediction tasks. In both datasets, adapting the off-the-shelf $FM_{SM}$ matched the performance of GBM models locally trained on all data while providing a 13% improvement in settings with few task-specific training labels. With continued pretraining on local data, label efficiency substantially improved, such that $FM_{SM}$ required fewer than 1% of training examples to match the fully trained GBM's performance. Continued pretraining was also 60 to 90% more sample-efficient than training local foundation models from scratch. Our findings show that adapting shared EHR foundation models across hospitals provides improved prediction performance at less cost, underscoring the utility of base foundation models as modular components to streamline the development of healthcare AI.
INSPECT: A Multimodal Dataset for Pulmonary Embolism Diagnosis and Prognosis
Nov 17, 2023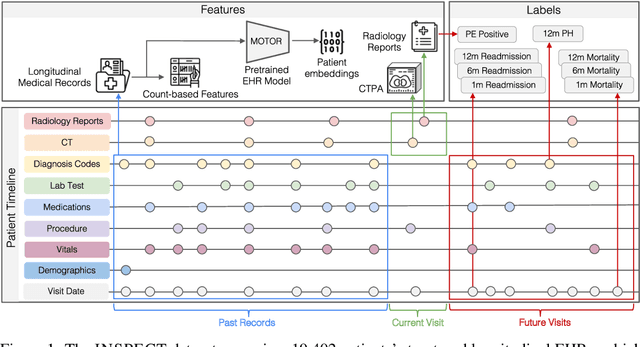
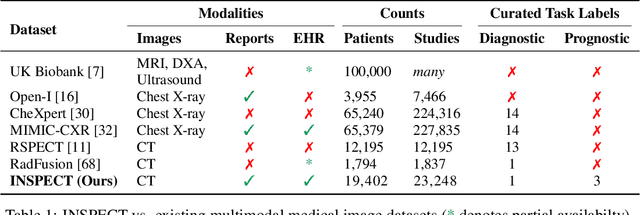
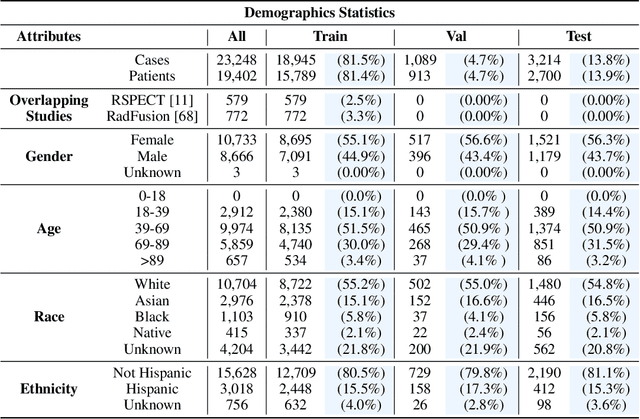
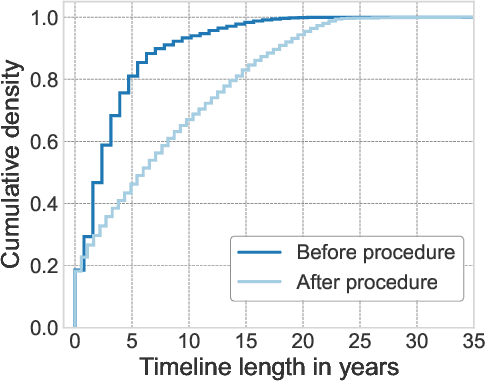
Abstract:Synthesizing information from multiple data sources plays a crucial role in the practice of modern medicine. Current applications of artificial intelligence in medicine often focus on single-modality data due to a lack of publicly available, multimodal medical datasets. To address this limitation, we introduce INSPECT, which contains de-identified longitudinal records from a large cohort of patients at risk for pulmonary embolism (PE), along with ground truth labels for multiple outcomes. INSPECT contains data from 19,402 patients, including CT images, radiology report impression sections, and structured electronic health record (EHR) data (i.e. demographics, diagnoses, procedures, vitals, and medications). Using INSPECT, we develop and release a benchmark for evaluating several baseline modeling approaches on a variety of important PE related tasks. We evaluate image-only, EHR-only, and multimodal fusion models. Trained models and the de-identified dataset are made available for non-commercial use under a data use agreement. To the best of our knowledge, INSPECT is the largest multimodal dataset integrating 3D medical imaging and EHR for reproducible methods evaluation and research.
MedAlign: A Clinician-Generated Dataset for Instruction Following with Electronic Medical Records
Aug 27, 2023
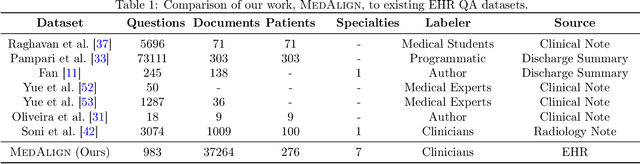
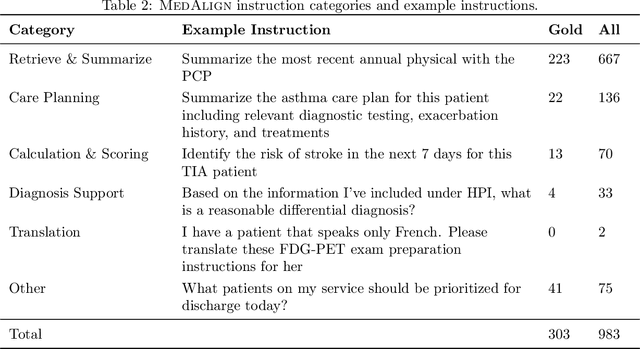

Abstract:The ability of large language models (LLMs) to follow natural language instructions with human-level fluency suggests many opportunities in healthcare to reduce administrative burden and improve quality of care. However, evaluating LLMs on realistic text generation tasks for healthcare remains challenging. Existing question answering datasets for electronic health record (EHR) data fail to capture the complexity of information needs and documentation burdens experienced by clinicians. To address these challenges, we introduce MedAlign, a benchmark dataset of 983 natural language instructions for EHR data. MedAlign is curated by 15 clinicians (7 specialities), includes clinician-written reference responses for 303 instructions, and provides 276 longitudinal EHRs for grounding instruction-response pairs. We used MedAlign to evaluate 6 general domain LLMs, having clinicians rank the accuracy and quality of each LLM response. We found high error rates, ranging from 35% (GPT-4) to 68% (MPT-7B-Instruct), and an 8.3% drop in accuracy moving from 32k to 2k context lengths for GPT-4. Finally, we report correlations between clinician rankings and automated natural language generation metrics as a way to rank LLMs without human review. We make MedAlign available under a research data use agreement to enable LLM evaluations on tasks aligned with clinician needs and preferences.
EHRSHOT: An EHR Benchmark for Few-Shot Evaluation of Foundation Models
Jul 05, 2023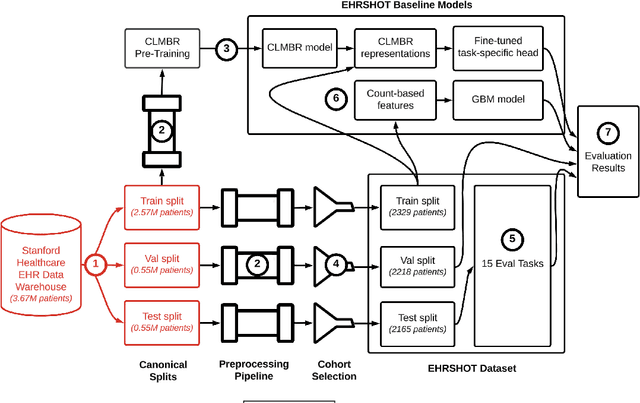
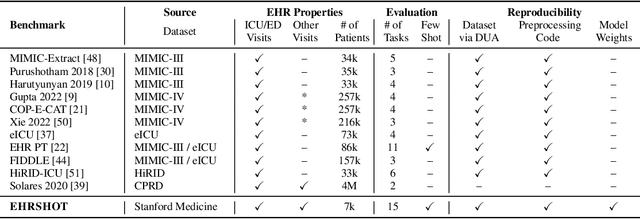

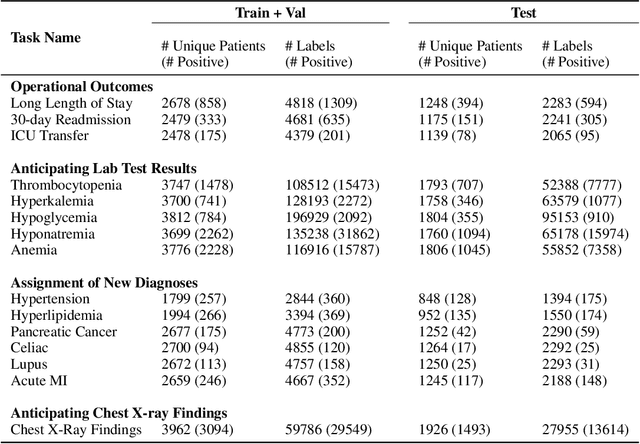
Abstract:While the general machine learning (ML) community has benefited from public datasets, tasks, and models, the progress of ML in healthcare has been hampered by a lack of such shared assets. The success of foundation models creates new challenges for healthcare ML by requiring access to shared pretrained models to validate performance benefits. We help address these challenges through three contributions. First, we publish a new dataset, EHRSHOT, containing de-identified structured data from the electronic health records (EHRs) of 6,712 patients from Stanford Medicine. Unlike MIMIC-III/IV and other popular EHR datasets, EHRSHOT is longitudinal and not restricted to ICU/ED patients. Second, we publish the weights of a 141M parameter clinical foundation model pretrained on the structured EHR data of 2.57M patients. We are one of the first to fully release such a model for coded EHR data; in contrast, most prior models released for clinical data (e.g. GatorTron, ClinicalBERT) only work with unstructured text and cannot process the rich, structured data within an EHR. We provide an end-to-end pipeline for the community to validate and build upon its performance. Third, we define 15 few-shot clinical prediction tasks, enabling evaluation of foundation models on benefits such as sample efficiency and task adaption. The code to reproduce our results, as well as the model and dataset (via a research data use agreement), are available at our Github repo here: https://github.com/som-shahlab/ehrshot-benchmark
The Shaky Foundations of Clinical Foundation Models: A Survey of Large Language Models and Foundation Models for EMRs
Mar 24, 2023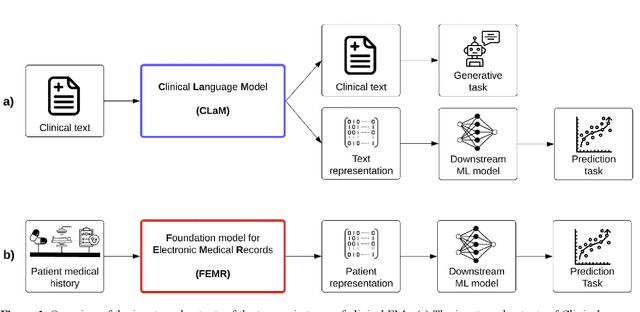
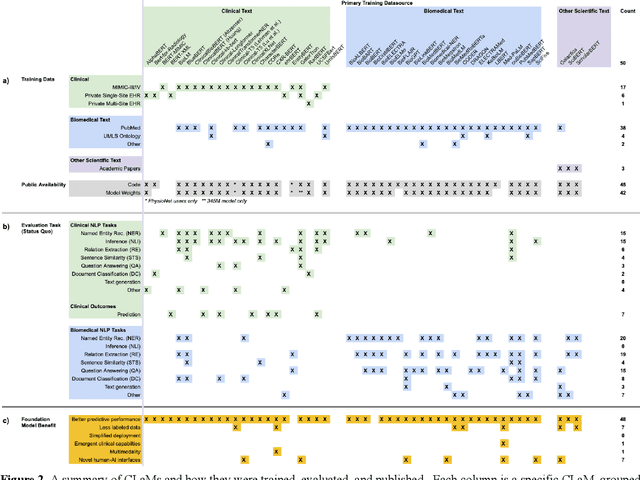
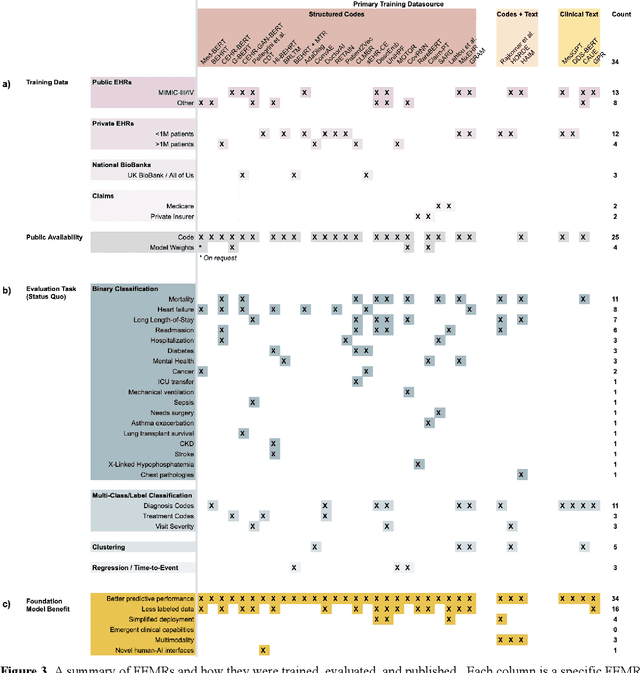

Abstract:The successes of foundation models such as ChatGPT and AlphaFold have spurred significant interest in building similar models for electronic medical records (EMRs) to improve patient care and hospital operations. However, recent hype has obscured critical gaps in our understanding of these models' capabilities. We review over 80 foundation models trained on non-imaging EMR data (i.e. clinical text and/or structured data) and create a taxonomy delineating their architectures, training data, and potential use cases. We find that most models are trained on small, narrowly-scoped clinical datasets (e.g. MIMIC-III) or broad, public biomedical corpora (e.g. PubMed) and are evaluated on tasks that do not provide meaningful insights on their usefulness to health systems. In light of these findings, we propose an improved evaluation framework for measuring the benefits of clinical foundation models that is more closely grounded to metrics that matter in healthcare.
Self-Supervised Time-to-Event Modeling with Structured Medical Records
Jan 09, 2023



Abstract:Time-to-event models (also known as survival models) are used in medicine and other fields for estimating the probability distribution of the time until a particular event occurs. While providing many advantages over traditional classification models, such as naturally handling censoring, time-to-event models require more parameters and are challenging to learn in settings with limited labeled training data. High censoring rates, common in events with long time horizons, further limit available training data and exacerbate the risk of overfitting. Existing methods, such as proportional hazard or accelerated failure time-based approaches, employ distributional assumptions to reduce parameter size, but they are vulnerable to model misspecification. In this work, we address these challenges with MOTOR, a self-supervised model that leverages temporal structure found in large-scale collections of timestamped, but largely unlabeled events, typical of electronic health record data. MOTOR defines a time-to-event pretraining task that naturally captures the probability distribution of event times, making it well-suited to applications in medicine. After pretraining on 8,192 tasks auto-generated from 2.7M patients (2.4B clinical events), we evaluate the performance of our pretrained model after fine-tuning to unseen time-to-event tasks. MOTOR-derived models improve upon current state-of-the-art C statistic performance by 6.6% and decrease training time (in wall time) by up to 8.2 times. We further improve sample efficiency, with adapted models matching current state-of-the-art performance using 95% less training data.
Instability in clinical risk stratification models using deep learning
Nov 20, 2022



Abstract:While it has been well known in the ML community that deep learning models suffer from instability, the consequences for healthcare deployments are under characterised. We study the stability of different model architectures trained on electronic health records, using a set of outpatient prediction tasks as a case study. We show that repeated training runs of the same deep learning model on the same training data can result in significantly different outcomes at a patient level even though global performance metrics remain stable. We propose two stability metrics for measuring the effect of randomness of model training, as well as mitigation strategies for improving model stability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge