Curtis Langlotz
RadDiff: Describing Differences in Radiology Image Sets with Natural Language
Jan 07, 2026Abstract:Understanding how two radiology image sets differ is critical for generating clinical insights and for interpreting medical AI systems. We introduce RadDiff, a multimodal agentic system that performs radiologist-style comparative reasoning to describe clinically meaningful differences between paired radiology studies. RadDiff builds on a proposer-ranker framework from VisDiff, and incorporates four innovations inspired by real diagnostic workflows: (1) medical knowledge injection through domain-adapted vision-language models; (2) multimodal reasoning that integrates images with their clinical reports; (3) iterative hypothesis refinement across multiple reasoning rounds; and (4) targeted visual search that localizes and zooms in on salient regions to capture subtle findings. To evaluate RadDiff, we construct RadDiffBench, a challenging benchmark comprising 57 expert-validated radiology study pairs with ground-truth difference descriptions. On RadDiffBench, RadDiff achieves 47% accuracy, and 50% accuracy when guided by ground-truth reports, significantly outperforming the general-domain VisDiff baseline. We further demonstrate RadDiff's versatility across diverse clinical tasks, including COVID-19 phenotype comparison, racial subgroup analysis, and discovery of survival-related imaging features. Together, RadDiff and RadDiffBench provide the first method-and-benchmark foundation for systematically uncovering meaningful differences in radiological data.
Improving the Performance of Radiology Report De-identification with Large-Scale Training and Benchmarking Against Cloud Vendor Methods
Nov 06, 2025



Abstract:Objective: To enhance automated de-identification of radiology reports by scaling transformer-based models through extensive training datasets and benchmarking performance against commercial cloud vendor systems for protected health information (PHI) detection. Materials and Methods: In this retrospective study, we built upon a state-of-the-art, transformer-based, PHI de-identification pipeline by fine-tuning on two large annotated radiology corpora from Stanford University, encompassing chest X-ray, chest CT, abdomen/pelvis CT, and brain MR reports and introducing an additional PHI category (AGE) into the architecture. Model performance was evaluated on test sets from Stanford and the University of Pennsylvania (Penn) for token-level PHI detection. We further assessed (1) the stability of synthetic PHI generation using a "hide-in-plain-sight" method and (2) performance against commercial systems. Precision, recall, and F1 scores were computed across all PHI categories. Results: Our model achieved overall F1 scores of 0.973 on the Penn dataset and 0.996 on the Stanford dataset, outperforming or maintaining the previous state-of-the-art model performance. Synthetic PHI evaluation showed consistent detectability (overall F1: 0.959 [0.958-0.960]) across 50 independently de-identified Penn datasets. Our model outperformed all vendor systems on synthetic Penn reports (overall F1: 0.960 vs. 0.632-0.754). Discussion: Large-scale, multimodal training improved cross-institutional generalization and robustness. Synthetic PHI generation preserved data utility while ensuring privacy. Conclusion: A transformer-based de-identification model trained on diverse radiology datasets outperforms prior academic and commercial systems in PHI detection and establishes a new benchmark for secure clinical text processing.
Automated Structured Radiology Report Generation
May 30, 2025Abstract:Automated radiology report generation from chest X-ray (CXR) images has the potential to improve clinical efficiency and reduce radiologists' workload. However, most datasets, including the publicly available MIMIC-CXR and CheXpert Plus, consist entirely of free-form reports, which are inherently variable and unstructured. This variability poses challenges for both generation and evaluation: existing models struggle to produce consistent, clinically meaningful reports, and standard evaluation metrics fail to capture the nuances of radiological interpretation. To address this, we introduce Structured Radiology Report Generation (SRRG), a new task that reformulates free-text radiology reports into a standardized format, ensuring clarity, consistency, and structured clinical reporting. We create a novel dataset by restructuring reports using large language models (LLMs) following strict structured reporting desiderata. Additionally, we introduce SRR-BERT, a fine-grained disease classification model trained on 55 labels, enabling more precise and clinically informed evaluation of structured reports. To assess report quality, we propose F1-SRR-BERT, a metric that leverages SRR-BERT's hierarchical disease taxonomy to bridge the gap between free-text variability and structured clinical reporting. We validate our dataset through a reader study conducted by five board-certified radiologists and extensive benchmarking experiments.
MedVAE: Efficient Automated Interpretation of Medical Images with Large-Scale Generalizable Autoencoders
Feb 20, 2025



Abstract:Medical images are acquired at high resolutions with large fields of view in order to capture fine-grained features necessary for clinical decision-making. Consequently, training deep learning models on medical images can incur large computational costs. In this work, we address the challenge of downsizing medical images in order to improve downstream computational efficiency while preserving clinically-relevant features. We introduce MedVAE, a family of six large-scale 2D and 3D autoencoders capable of encoding medical images as downsized latent representations and decoding latent representations back to high-resolution images. We train MedVAE autoencoders using a novel two-stage training approach with 1,052,730 medical images. Across diverse tasks obtained from 20 medical image datasets, we demonstrate that (1) utilizing MedVAE latent representations in place of high-resolution images when training downstream models can lead to efficiency benefits (up to 70x improvement in throughput) while simultaneously preserving clinically-relevant features and (2) MedVAE can decode latent representations back to high-resolution images with high fidelity. Our work demonstrates that large-scale, generalizable autoencoders can help address critical efficiency challenges in the medical domain. Our code is available at https://github.com/StanfordMIMI/MedVAE.
Best Practices for Large Language Models in Radiology
Dec 02, 2024



Abstract:At the heart of radiological practice is the challenge of integrating complex imaging data with clinical information to produce actionable insights. Nuanced application of language is key for various activities, including managing requests, describing and interpreting imaging findings in the context of clinical data, and concisely documenting and communicating the outcomes. The emergence of large language models (LLMs) offers an opportunity to improve the management and interpretation of the vast data in radiology. Despite being primarily general-purpose, these advanced computational models demonstrate impressive capabilities in specialized language-related tasks, even without specific training. Unlocking the potential of LLMs for radiology requires basic understanding of their foundations and a strategic approach to navigate their idiosyncrasies. This review, drawing from practical radiology and machine learning expertise and recent literature, provides readers insight into the potential of LLMs in radiology. It examines best practices that have so far stood the test of time in the rapidly evolving landscape of LLMs. This includes practical advice for optimizing LLM characteristics for radiology practices along with limitations, effective prompting, and fine-tuning strategies.
Foundation Models in Radiology: What, How, When, Why and Why Not
Nov 27, 2024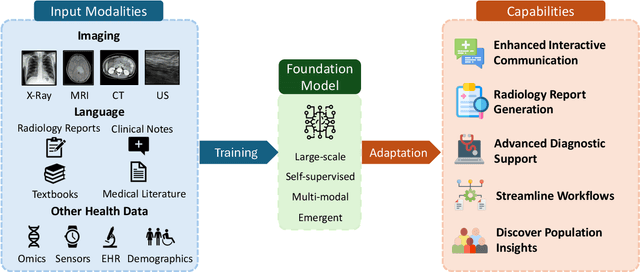
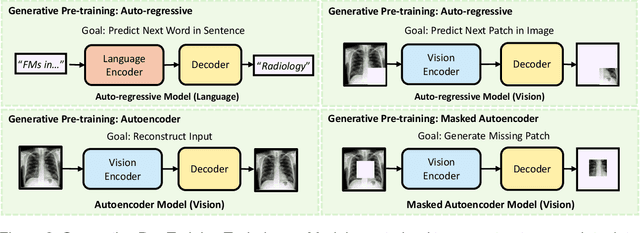
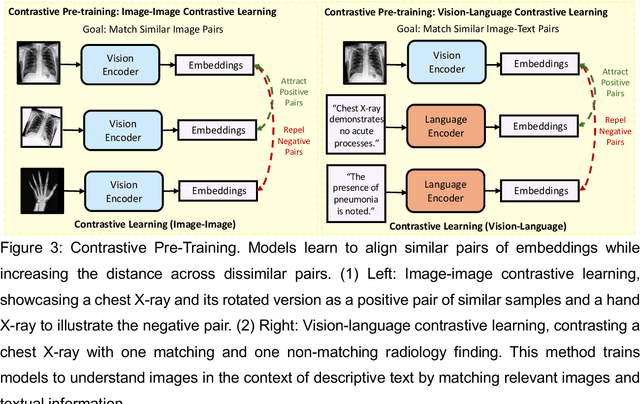
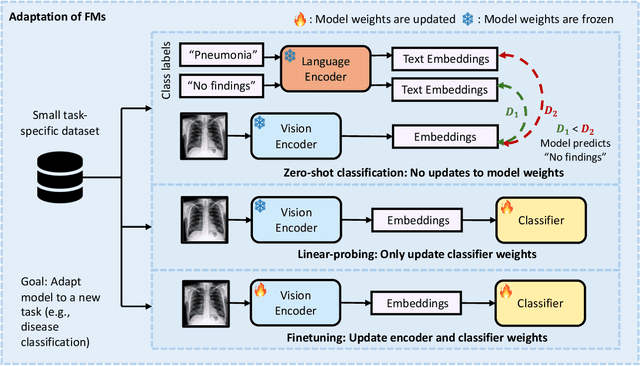
Abstract:Recent advances in artificial intelligence have witnessed the emergence of large-scale deep learning models capable of interpreting and generating both textual and imaging data. Such models, typically referred to as foundation models, are trained on extensive corpora of unlabeled data and demonstrate high performance across various tasks. Foundation models have recently received extensive attention from academic, industry, and regulatory bodies. Given the potentially transformative impact that foundation models can have on the field of radiology, this review aims to establish a standardized terminology concerning foundation models, with a specific focus on the requirements of training data, model training paradigms, model capabilities, and evaluation strategies. We further outline potential pathways to facilitate the training of radiology-specific foundation models, with a critical emphasis on elucidating both the benefits and challenges associated with such models. Overall, we envision that this review can unify technical advances and clinical needs in the training of foundation models for radiology in a safe and responsible manner, for ultimately benefiting patients, providers, and radiologists.
Evaluating and Improving the Effectiveness of Synthetic Chest X-Rays for Medical Image Analysis
Nov 27, 2024Abstract:Purpose: To explore best-practice approaches for generating synthetic chest X-ray images and augmenting medical imaging datasets to optimize the performance of deep learning models in downstream tasks like classification and segmentation. Materials and Methods: We utilized a latent diffusion model to condition the generation of synthetic chest X-rays on text prompts and/or segmentation masks. We explored methods like using a proxy model and using radiologist feedback to improve the quality of synthetic data. These synthetic images were then generated from relevant disease information or geometrically transformed segmentation masks and added to ground truth training set images from the CheXpert, CANDID-PTX, SIIM, and RSNA Pneumonia datasets to measure improvements in classification and segmentation model performance on the test sets. F1 and Dice scores were used to evaluate classification and segmentation respectively. One-tailed t-tests with Bonferroni correction assessed the statistical significance of performance improvements with synthetic data. Results: Across all experiments, the synthetic data we generated resulted in a maximum mean classification F1 score improvement of 0.150453 (CI: 0.099108-0.201798; P=0.0031) compared to using only real data. For segmentation, the maximum Dice score improvement was 0.14575 (CI: 0.108267-0.183233; P=0.0064). Conclusion: Best practices for generating synthetic chest X-ray images for downstream tasks include conditioning on single-disease labels or geometrically transformed segmentation masks, as well as potentially using proxy modeling for fine-tuning such generations.
Time-to-Event Pretraining for 3D Medical Imaging
Nov 14, 2024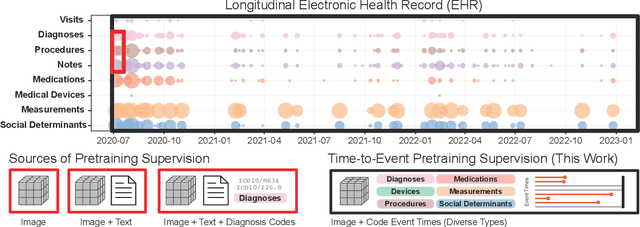
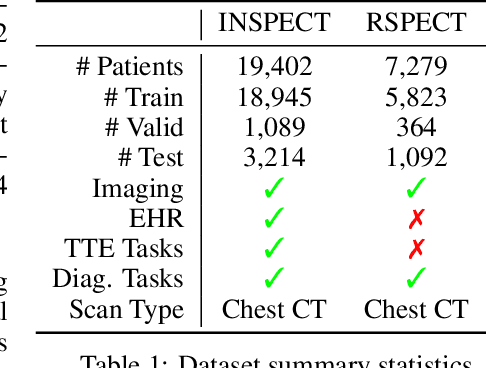
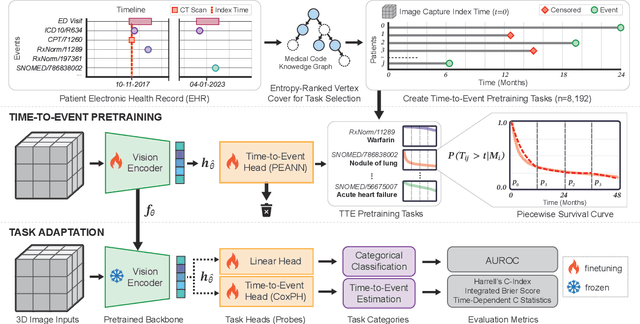
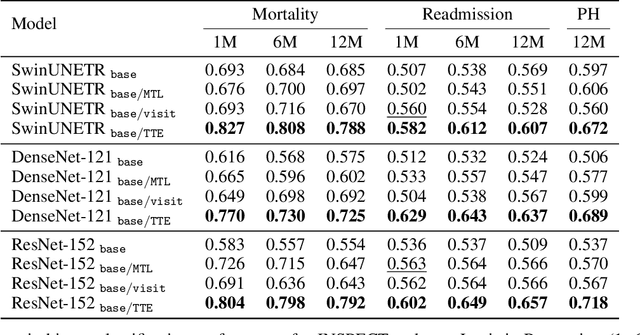
Abstract:With the rise of medical foundation models and the growing availability of imaging data, scalable pretraining techniques offer a promising way to identify imaging biomarkers predictive of future disease risk. While current self-supervised methods for 3D medical imaging models capture local structural features like organ morphology, they fail to link pixel biomarkers with long-term health outcomes due to a missing context problem. Current approaches lack the temporal context necessary to identify biomarkers correlated with disease progression, as they rely on supervision derived only from images and concurrent text descriptions. To address this, we introduce time-to-event pretraining, a pretraining framework for 3D medical imaging models that leverages large-scale temporal supervision from paired, longitudinal electronic health records (EHRs). Using a dataset of 18,945 CT scans (4.2 million 2D images) and time-to-event distributions across thousands of EHR-derived tasks, our method improves outcome prediction, achieving an average AUROC increase of 23.7% and a 29.4% gain in Harrell's C-index across 8 benchmark tasks. Importantly, these gains are achieved without sacrificing diagnostic classification performance. This study lays the foundation for integrating longitudinal EHR and 3D imaging data to advance clinical risk prediction.
RaVL: Discovering and Mitigating Spurious Correlations in Fine-Tuned Vision-Language Models
Nov 06, 2024



Abstract:Fine-tuned vision-language models (VLMs) often capture spurious correlations between image features and textual attributes, resulting in degraded zero-shot performance at test time. Existing approaches for addressing spurious correlations (i) primarily operate at the global image-level rather than intervening directly on fine-grained image features and (ii) are predominantly designed for unimodal settings. In this work, we present RaVL, which takes a fine-grained perspective on VLM robustness by discovering and mitigating spurious correlations using local image features rather than operating at the global image level. Given a fine-tuned VLM, RaVL first discovers spurious correlations by leveraging a region-level clustering approach to identify precise image features contributing to zero-shot classification errors. Then, RaVL mitigates the identified spurious correlation with a novel region-aware loss function that enables the VLM to focus on relevant regions and ignore spurious relationships during fine-tuning. We evaluate RaVL on 654 VLMs with various model architectures, data domains, and learned spurious correlations. Our results show that RaVL accurately discovers (191% improvement over the closest baseline) and mitigates (8.2% improvement on worst-group image classification accuracy) spurious correlations. Qualitative evaluations on general-domain and medical-domain VLMs confirm our findings.
Preference Fine-Tuning for Factuality in Chest X-Ray Interpretation Models Without Human Feedback
Oct 09, 2024



Abstract:Radiologists play a crucial role by translating medical images into medical reports. However, the field faces staffing shortages and increasing workloads. While automated approaches using vision-language models (VLMs) show promise as assistants, they require exceptionally high accuracy. Most current VLMs in radiology rely solely on supervised fine-tuning (SFT). Meanwhile, in the general domain, additional preference fine-tuning has become standard practice. The challenge in radiology lies in the prohibitive cost of obtaining radiologist feedback. We propose a scalable automated preference alignment technique for VLMs in radiology, focusing on chest X-ray (CXR) report generation. Our method leverages publicly available datasets with an LLM-as-a-Judge mechanism, eliminating the need for additional expert radiologist feedback. We evaluate and benchmark five direct alignment algorithms (DAAs). Our results show up to a 57.4% improvement in average GREEN scores, a LLM-based metric for evaluating CXR reports, and a 9.2% increase in an average across six metrics (domain specific and general), compared to the SFT baseline. We study reward overoptimization via length exploitation, with reports lengthening by up to 3.2x. To assess a potential alignment tax, we benchmark on six additional diverse tasks, finding no significant degradations. A reader study involving four board-certified radiologists indicates win rates of up to 0.62 over the SFT baseline, while significantly penalizing verbosity. Our analysis provides actionable insights for the development of VLMs in high-stakes fields like radiology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge