Nigam H. Shah
Atropos Health, New York NY, USA, Center for Biomedical Informatics Research, Stanford University, Stanford CA, USA
Monitoring Deployed AI Systems in Health Care
Dec 09, 2025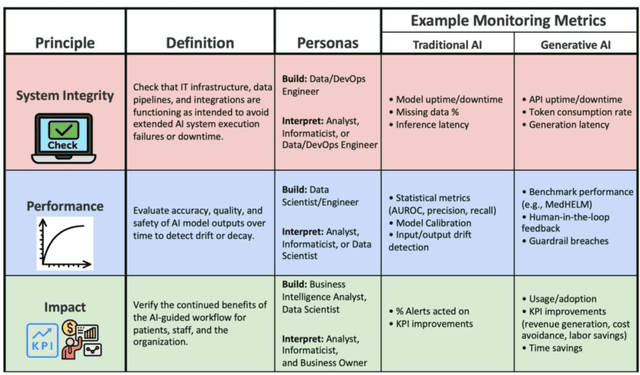
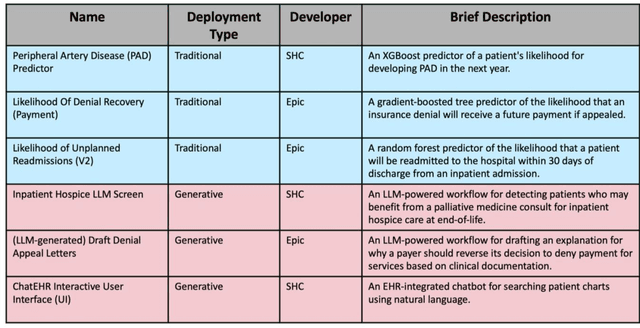
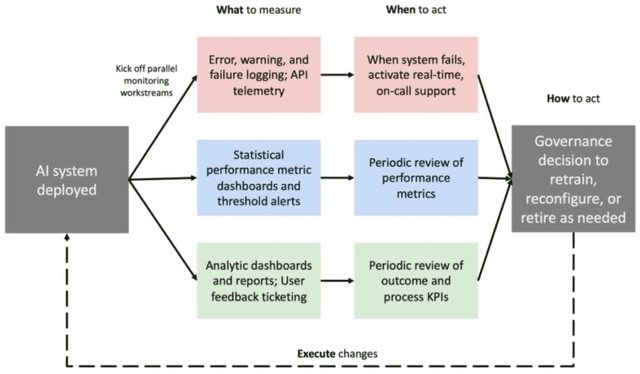
Abstract:Post-deployment monitoring of artificial intelligence (AI) systems in health care is essential to ensure their safety, quality, and sustained benefit-and to support governance decisions about which systems to update, modify, or decommission. Motivated by these needs, we developed a framework for monitoring deployed AI systems grounded in the mandate to take specific actions when they fail to behave as intended. This framework, which is now actively used at Stanford Health Care, is organized around three complementary principles: system integrity, performance, and impact. System integrity monitoring focuses on maximizing system uptime, detecting runtime errors, and identifying when changes to the surrounding IT ecosystem have unintended effects. Performance monitoring focuses on maintaining accurate system behavior in the face of changing health care practices (and thus input data) over time. Impact monitoring assesses whether a deployed system continues to have value in the form of benefit to clinicians and patients. Drawing on examples of deployed AI systems at our academic medical center, we provide practical guidance for creating monitoring plans based on these principles that specify which metrics to measure, when those metrics should be reviewed, who is responsible for acting when metrics change, and what concrete follow-up actions should be taken-for both traditional and generative AI. We also discuss challenges to implementing this framework, including the effort and cost of monitoring for health systems with limited resources and the difficulty of incorporating data-driven monitoring practices into complex organizations where conflicting priorities and definitions of success often coexist. This framework offers a practical template and starting point for health systems seeking to ensure that AI deployments remain safe and effective over time.
The Optimization Paradox in Clinical AI Multi-Agent Systems
Jun 06, 2025Abstract:Multi-agent artificial intelligence systems are increasingly deployed in clinical settings, yet the relationship between component-level optimization and system-wide performance remains poorly understood. We evaluated this relationship using 2,400 real patient cases from the MIMIC-CDM dataset across four abdominal pathologies (appendicitis, pancreatitis, cholecystitis, diverticulitis), decomposing clinical diagnosis into information gathering, interpretation, and differential diagnosis. We evaluated single agent systems (one model performing all tasks) against multi-agent systems (specialized models for each task) using comprehensive metrics spanning diagnostic outcomes, process adherence, and cost efficiency. Our results reveal a paradox: while multi-agent systems generally outperformed single agents, the component-optimized or Best of Breed system with superior components and excellent process metrics (85.5% information accuracy) significantly underperformed in diagnostic accuracy (67.7% vs. 77.4% for a top multi-agent system). This finding underscores that successful integration of AI in healthcare requires not just component level optimization but also attention to information flow and compatibility between agents. Our findings highlight the need for end to end system validation rather than relying on component metrics alone.
MedHELM: Holistic Evaluation of Large Language Models for Medical Tasks
May 26, 2025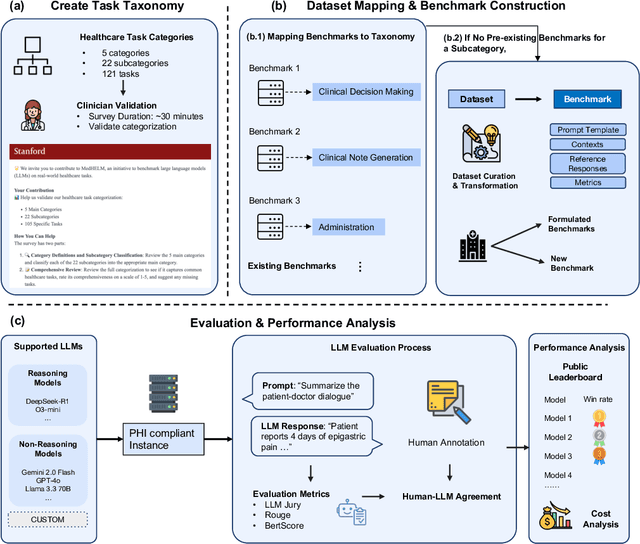
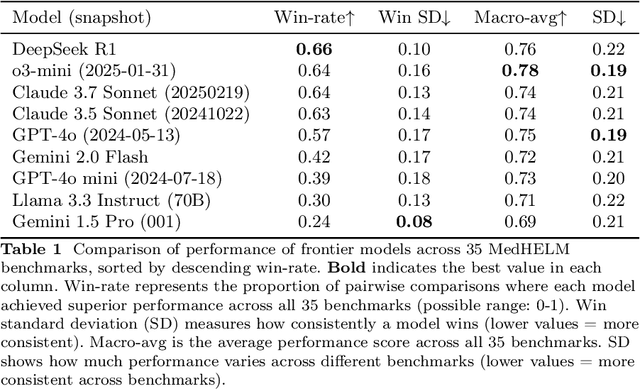
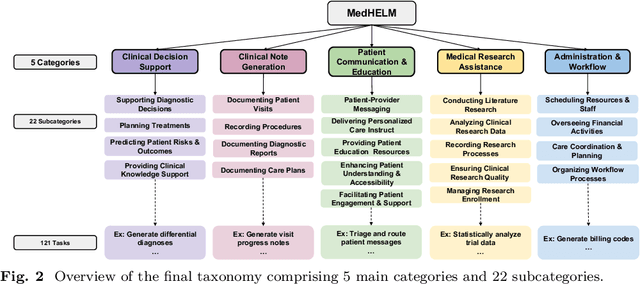
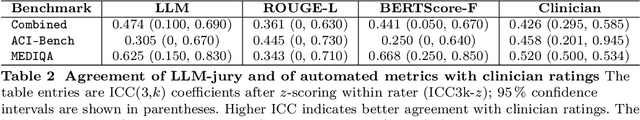
Abstract:While large language models (LLMs) achieve near-perfect scores on medical licensing exams, these evaluations inadequately reflect the complexity and diversity of real-world clinical practice. We introduce MedHELM, an extensible evaluation framework for assessing LLM performance for medical tasks with three key contributions. First, a clinician-validated taxonomy spanning 5 categories, 22 subcategories, and 121 tasks developed with 29 clinicians. Second, a comprehensive benchmark suite comprising 35 benchmarks (17 existing, 18 newly formulated) providing complete coverage of all categories and subcategories in the taxonomy. Third, a systematic comparison of LLMs with improved evaluation methods (using an LLM-jury) and a cost-performance analysis. Evaluation of 9 frontier LLMs, using the 35 benchmarks, revealed significant performance variation. Advanced reasoning models (DeepSeek R1: 66% win-rate; o3-mini: 64% win-rate) demonstrated superior performance, though Claude 3.5 Sonnet achieved comparable results at 40% lower estimated computational cost. On a normalized accuracy scale (0-1), most models performed strongly in Clinical Note Generation (0.73-0.85) and Patient Communication & Education (0.78-0.83), moderately in Medical Research Assistance (0.65-0.75), and generally lower in Clinical Decision Support (0.56-0.72) and Administration & Workflow (0.53-0.63). Our LLM-jury evaluation method achieved good agreement with clinician ratings (ICC = 0.47), surpassing both average clinician-clinician agreement (ICC = 0.43) and automated baselines including ROUGE-L (0.36) and BERTScore-F1 (0.44). Claude 3.5 Sonnet achieved comparable performance to top models at lower estimated cost. These findings highlight the importance of real-world, task-specific evaluation for medical use of LLMs and provides an open source framework to enable this.
Context Clues: Evaluating Long Context Models for Clinical Prediction Tasks on EHRs
Dec 09, 2024



Abstract:Foundation Models (FMs) trained on Electronic Health Records (EHRs) have achieved state-of-the-art results on numerous clinical prediction tasks. However, most existing EHR FMs have context windows of <1k tokens. This prevents them from modeling full patient EHRs which can exceed 10k's of events. Recent advancements in subquadratic long-context architectures (e.g., Mamba) offer a promising solution. However, their application to EHR data has not been well-studied. We address this gap by presenting the first systematic evaluation of the effect of context length on modeling EHR data. We find that longer context models improve predictive performance -- our Mamba-based model surpasses the prior state-of-the-art on 9/14 tasks on the EHRSHOT prediction benchmark. For clinical applications, however, model performance alone is insufficient -- robustness to the unique properties of EHR is crucial. Thus, we also evaluate models across three previously underexplored properties of EHR data: (1) the prevalence of "copy-forwarded" diagnoses which creates artificial repetition of tokens within EHR sequences; (2) the irregular time intervals between EHR events which can lead to a wide range of timespans within a context window; and (3) the natural increase in disease complexity over time which makes later tokens in the EHR harder to predict than earlier ones. Stratifying our EHRSHOT results, we find that higher levels of each property correlate negatively with model performance, but that longer context models are more robust to more extreme levels of these properties. Our work highlights the potential for using long-context architectures to model EHR data, and offers a case study for identifying new challenges in modeling sequential data motivated by domains outside of natural language. We release our models and code at: https://github.com/som-shahlab/long_context_clues
Time-to-Event Pretraining for 3D Medical Imaging
Nov 14, 2024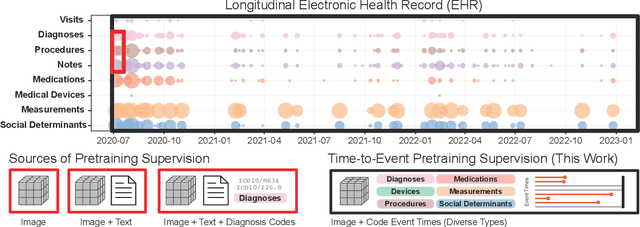
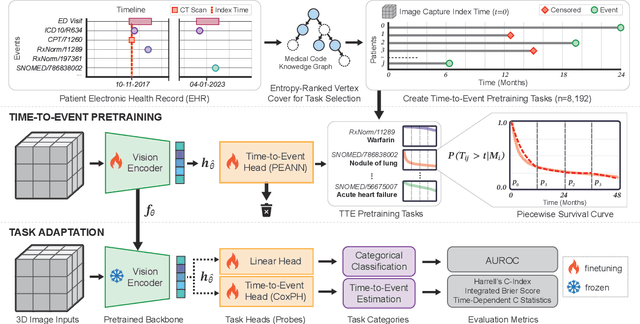
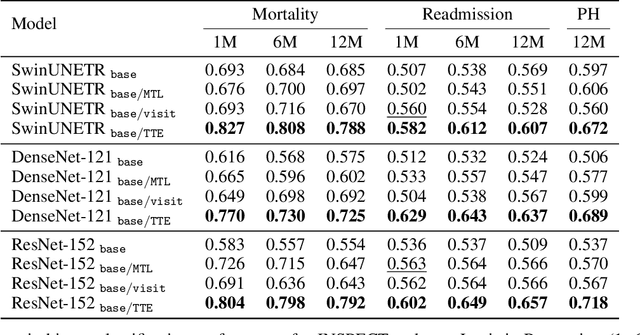
Abstract:With the rise of medical foundation models and the growing availability of imaging data, scalable pretraining techniques offer a promising way to identify imaging biomarkers predictive of future disease risk. While current self-supervised methods for 3D medical imaging models capture local structural features like organ morphology, they fail to link pixel biomarkers with long-term health outcomes due to a missing context problem. Current approaches lack the temporal context necessary to identify biomarkers correlated with disease progression, as they rely on supervision derived only from images and concurrent text descriptions. To address this, we introduce time-to-event pretraining, a pretraining framework for 3D medical imaging models that leverages large-scale temporal supervision from paired, longitudinal electronic health records (EHRs). Using a dataset of 18,945 CT scans (4.2 million 2D images) and time-to-event distributions across thousands of EHR-derived tasks, our method improves outcome prediction, achieving an average AUROC increase of 23.7% and a 29.4% gain in Harrell's C-index across 8 benchmark tasks. Importantly, these gains are achieved without sacrificing diagnostic classification performance. This study lays the foundation for integrating longitudinal EHR and 3D imaging data to advance clinical risk prediction.
meds_reader: A fast and efficient EHR processing library
Sep 12, 2024



Abstract:The growing demand for machine learning in healthcare requires processing increasingly large electronic health record (EHR) datasets, but existing pipelines are not computationally efficient or scalable. In this paper, we introduce meds_reader, an optimized Python package for efficient EHR data processing that is designed to take advantage of many intrinsic properties of EHR data for improved speed. We then demonstrate the benefits of meds_reader by reimplementing key components of two major EHR processing pipelines, achieving 10-100x improvements in memory, speed, and disk usage. The code for meds_reader can be found at https://github.com/som-shahlab/meds_reader.
Answering real-world clinical questions using large language model based systems
Jun 29, 2024



Abstract:Evidence to guide healthcare decisions is often limited by a lack of relevant and trustworthy literature as well as difficulty in contextualizing existing research for a specific patient. Large language models (LLMs) could potentially address both challenges by either summarizing published literature or generating new studies based on real-world data (RWD). We evaluated the ability of five LLM-based systems in answering 50 clinical questions and had nine independent physicians review the responses for relevance, reliability, and actionability. As it stands, general-purpose LLMs (ChatGPT-4, Claude 3 Opus, Gemini Pro 1.5) rarely produced answers that were deemed relevant and evidence-based (2% - 10%). In contrast, retrieval augmented generation (RAG)-based and agentic LLM systems produced relevant and evidence-based answers for 24% (OpenEvidence) to 58% (ChatRWD) of questions. Only the agentic ChatRWD was able to answer novel questions compared to other LLMs (65% vs. 0-9%). These results suggest that while general-purpose LLMs should not be used as-is, a purpose-built system for evidence summarization based on RAG and one for generating novel evidence working synergistically would improve availability of pertinent evidence for patient care.
Do Multimodal Foundation Models Understand Enterprise Workflows? A Benchmark for Business Process Management Tasks
Jun 19, 2024



Abstract:Existing ML benchmarks lack the depth and diversity of annotations needed for evaluating models on business process management (BPM) tasks. BPM is the practice of documenting, measuring, improving, and automating enterprise workflows. However, research has focused almost exclusively on one task - full end-to-end automation using agents based on multimodal foundation models (FMs) like GPT-4. This focus on automation ignores the reality of how most BPM tools are applied today - simply documenting the relevant workflow takes 60% of the time of the typical process optimization project. To address this gap we present WONDERBREAD, the first benchmark for evaluating multimodal FMs on BPM tasks beyond automation. Our contributions are: (1) a dataset containing 2928 documented workflow demonstrations; (2) 6 novel BPM tasks sourced from real-world applications ranging from workflow documentation to knowledge transfer to process improvement; and (3) an automated evaluation harness. Our benchmark shows that while state-of-the-art FMs can automatically generate documentation (e.g. recalling 88% of the steps taken in a video demonstration of a workflow), they struggle to re-apply that knowledge towards finer-grained validation of workflow completion (F1 < 0.3). We hope WONDERBREAD encourages the development of more "human-centered" AI tooling for enterprise applications and furthers the exploration of multimodal FMs for the broader universe of BPM tasks. We publish our dataset and experiments here: https://github.com/HazyResearch/wonderbread
Merlin: A Vision Language Foundation Model for 3D Computed Tomography
Jun 10, 2024



Abstract:Over 85 million computed tomography (CT) scans are performed annually in the US, of which approximately one quarter focus on the abdomen. Given the current radiologist shortage, there is a large impetus to use artificial intelligence to alleviate the burden of interpreting these complex imaging studies. Prior state-of-the-art approaches for automated medical image interpretation leverage vision language models (VLMs). However, current medical VLMs are generally limited to 2D images and short reports, and do not leverage electronic health record (EHR) data for supervision. We introduce Merlin - a 3D VLM that we train using paired CT scans (6+ million images from 15,331 CTs), EHR diagnosis codes (1.8+ million codes), and radiology reports (6+ million tokens). We evaluate Merlin on 6 task types and 752 individual tasks. The non-adapted (off-the-shelf) tasks include zero-shot findings classification (31 findings), phenotype classification (692 phenotypes), and zero-shot cross-modal retrieval (image to findings and image to impressions), while model adapted tasks include 5-year disease prediction (6 diseases), radiology report generation, and 3D semantic segmentation (20 organs). We perform internal validation on a test set of 5,137 CTs, and external validation on 7,000 clinical CTs and on two public CT datasets (VerSe, TotalSegmentator). Beyond these clinically-relevant evaluations, we assess the efficacy of various network architectures and training strategies to depict that Merlin has favorable performance to existing task-specific baselines. We derive data scaling laws to empirically assess training data needs for requisite downstream task performance. Furthermore, unlike conventional VLMs that require hundreds of GPUs for training, we perform all training on a single GPU.
Automating the Enterprise with Foundation Models
May 03, 2024



Abstract:Automating enterprise workflows could unlock $4 trillion/year in productivity gains. Despite being of interest to the data management community for decades, the ultimate vision of end-to-end workflow automation has remained elusive. Current solutions rely on process mining and robotic process automation (RPA), in which a bot is hard-coded to follow a set of predefined rules for completing a workflow. Through case studies of a hospital and large B2B enterprise, we find that the adoption of RPA has been inhibited by high set-up costs (12-18 months), unreliable execution (60% initial accuracy), and burdensome maintenance (requiring multiple FTEs). Multimodal foundation models (FMs) such as GPT-4 offer a promising new approach for end-to-end workflow automation given their generalized reasoning and planning abilities. To study these capabilities we propose ECLAIR, a system to automate enterprise workflows with minimal human supervision. We conduct initial experiments showing that multimodal FMs can address the limitations of traditional RPA with (1) near-human-level understanding of workflows (93% accuracy on a workflow understanding task) and (2) instant set-up with minimal technical barrier (based solely on a natural language description of a workflow, ECLAIR achieves end-to-end completion rates of 40%). We identify human-AI collaboration, validation, and self-improvement as open challenges, and suggest ways they can be solved with data management techniques. Code is available at: https://github.com/HazyResearch/eclair-agents
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge