Hejie Cui
HeaPA: Difficulty-Aware Heap Sampling and On-Policy Query Augmentation for LLM Reinforcement Learning
Jan 30, 2026Abstract:RLVR is now a standard way to train LLMs on reasoning tasks with verifiable outcomes, but when rollout generation dominates the cost, efficiency depends heavily on which prompts you sample and when. In practice, prompt pools are often static or only loosely tied to the model's learning progress, so uniform sampling can't keep up with the shifting capability frontier and ends up wasting rollouts on prompts that are already solved or still out of reach. Existing approaches improve efficiency through filtering, curricula, adaptive rollout allocation, or teacher guidance, but they typically assume a fixed pool-which makes it hard to support stable on-policy pool growth-or they add extra teacher cost and latency. We introduce HeaPA (Heap Sampling and On-Policy Query Augmentation), which maintains a bounded, evolving pool, tracks the frontier using heap-based boundary sampling, expands the pool via on-policy augmentation with lightweight asynchronous validation, and stabilizes correlated queries through topology-aware re-estimation of pool statistics and controlled reinsertion. Across two training corpora, two training recipes, and seven benchmarks, HeaPA consistently improves accuracy and reaches target performance with fewer computations while keeping wall-clock time comparable. Our analyses suggest these gains come from frontier-focused sampling and on-policy pool growth, with the benefits becoming larger as model scale increases. Our code is available at https://github.com/horizon-rl/HeaPA.
WebCoach: Self-Evolving Web Agents with Cross-Session Memory Guidance
Nov 17, 2025Abstract:Multimodal LLM-powered agents have recently demonstrated impressive capabilities in web navigation, enabling agents to complete complex browsing tasks across diverse domains. However, current agents struggle with repetitive errors and lack the ability to learn from past experiences across sessions, limiting their long-term robustness and sample efficiency. We introduce WebCoach, a model-agnostic self-evolving framework that equips web browsing agents with persistent cross-session memory, enabling improved long-term planning, reflection, and continual learning without retraining. WebCoach consists of three key components: (1) a WebCondenser, which standardizes raw navigation logs into concise summaries; (2) an External Memory Store, which organizes complete trajectories as episodic experiences; and (3) a Coach, which retrieves relevant experiences based on similarity and recency, and decides whether to inject task-specific advice into the agent via runtime hooks. This design empowers web agents to access long-term memory beyond their native context window, improving robustness in complex browsing tasks. Moreover, WebCoach achieves self-evolution by continuously curating episodic memory from new navigation trajectories, enabling agents to improve over time without retraining. Evaluations on the WebVoyager benchmark demonstrate that WebCoach consistently improves the performance of browser-use agents across three different LLM backbones. With a 38B model, it increases task success rates from 47% to 61% while reducing or maintaining the average number of steps. Notably, smaller base models with WebCoach achieve performance comparable to the same web agent using GPT-4o.
KERAP: A Knowledge-Enhanced Reasoning Approach for Accurate Zero-shot Diagnosis Prediction Using Multi-agent LLMs
Jul 03, 2025



Abstract:Medical diagnosis prediction plays a critical role in disease detection and personalized healthcare. While machine learning (ML) models have been widely adopted for this task, their reliance on supervised training limits their ability to generalize to unseen cases, particularly given the high cost of acquiring large, labeled datasets. Large language models (LLMs) have shown promise in leveraging language abilities and biomedical knowledge for diagnosis prediction. However, they often suffer from hallucinations, lack structured medical reasoning, and produce useless outputs. To address these challenges, we propose KERAP, a knowledge graph (KG)-enhanced reasoning approach that improves LLM-based diagnosis prediction through a multi-agent architecture. Our framework consists of a linkage agent for attribute mapping, a retrieval agent for structured knowledge extraction, and a prediction agent that iteratively refines diagnosis predictions. Experimental results demonstrate that KERAP enhances diagnostic reliability efficiently, offering a scalable and interpretable solution for zero-shot medical diagnosis prediction.
MedHELM: Holistic Evaluation of Large Language Models for Medical Tasks
May 26, 2025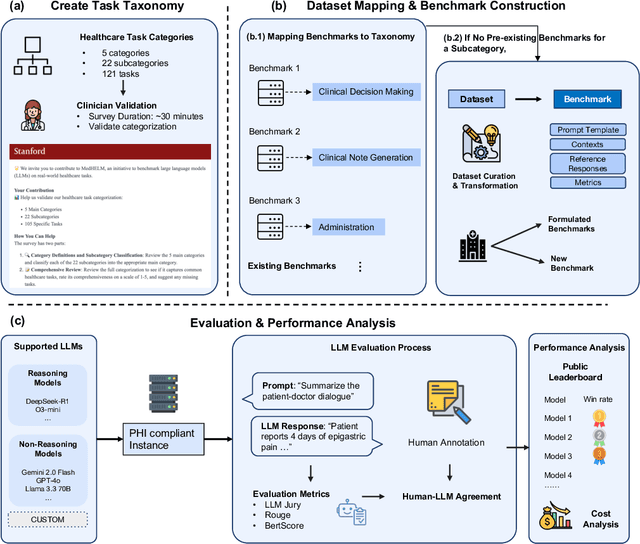
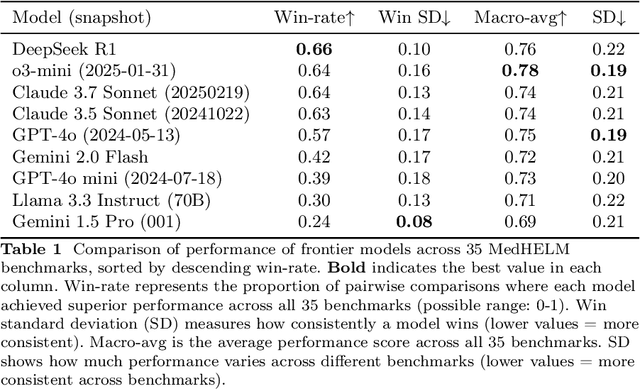
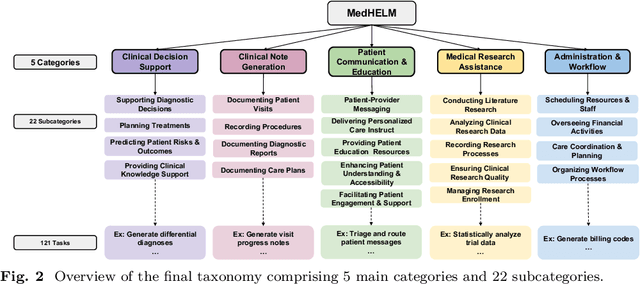
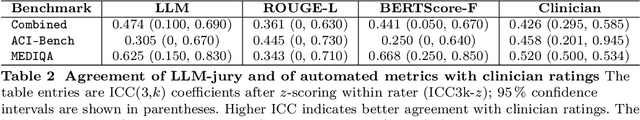
Abstract:While large language models (LLMs) achieve near-perfect scores on medical licensing exams, these evaluations inadequately reflect the complexity and diversity of real-world clinical practice. We introduce MedHELM, an extensible evaluation framework for assessing LLM performance for medical tasks with three key contributions. First, a clinician-validated taxonomy spanning 5 categories, 22 subcategories, and 121 tasks developed with 29 clinicians. Second, a comprehensive benchmark suite comprising 35 benchmarks (17 existing, 18 newly formulated) providing complete coverage of all categories and subcategories in the taxonomy. Third, a systematic comparison of LLMs with improved evaluation methods (using an LLM-jury) and a cost-performance analysis. Evaluation of 9 frontier LLMs, using the 35 benchmarks, revealed significant performance variation. Advanced reasoning models (DeepSeek R1: 66% win-rate; o3-mini: 64% win-rate) demonstrated superior performance, though Claude 3.5 Sonnet achieved comparable results at 40% lower estimated computational cost. On a normalized accuracy scale (0-1), most models performed strongly in Clinical Note Generation (0.73-0.85) and Patient Communication & Education (0.78-0.83), moderately in Medical Research Assistance (0.65-0.75), and generally lower in Clinical Decision Support (0.56-0.72) and Administration & Workflow (0.53-0.63). Our LLM-jury evaluation method achieved good agreement with clinician ratings (ICC = 0.47), surpassing both average clinician-clinician agreement (ICC = 0.43) and automated baselines including ROUGE-L (0.36) and BERTScore-F1 (0.44). Claude 3.5 Sonnet achieved comparable performance to top models at lower estimated cost. These findings highlight the importance of real-world, task-specific evaluation for medical use of LLMs and provides an open source framework to enable this.
Data Foundations for Large Scale Multimodal Clinical Foundation Models
Mar 09, 2025Abstract:Recent advances in clinical AI have enabled remarkable progress across many clinical domains. However, existing benchmarks and models are primarily limited to a small set of modalities and tasks, which hinders the development of large-scale multimodal methods that can make holistic assessments of patient health and well-being. To bridge this gap, we introduce Clinical Large-Scale Integrative Multimodal Benchmark (CLIMB), a comprehensive clinical benchmark unifying diverse clinical data across imaging, language, temporal, and graph modalities. CLIMB comprises 4.51 million patient samples totaling 19.01 terabytes distributed across 2D imaging, 3D video, time series, graphs, and multimodal data. Through extensive empirical evaluation, we demonstrate that multitask pretraining significantly improves performance on understudied domains, achieving up to 29% improvement in ultrasound and 23% in ECG analysis over single-task learning. Pretraining on CLIMB also effectively improves models' generalization capability to new tasks, and strong unimodal encoder performance translates well to multimodal performance when paired with task-appropriate fusion strategies. Our findings provide a foundation for new architecture designs and pretraining strategies to advance clinical AI research. Code is released at https://github.com/DDVD233/climb.
TIMER: Temporal Instruction Modeling and Evaluation for Longitudinal Clinical Records
Mar 06, 2025Abstract:Large language models (LLMs) have emerged as promising tools for assisting in medical tasks, yet processing Electronic Health Records (EHRs) presents unique challenges due to their longitudinal nature. While LLMs' capabilities to perform medical tasks continue to improve, their ability to reason over temporal dependencies across multiple patient visits and time frames remains unexplored. We introduce TIMER (Temporal Instruction Modeling and Evaluation for Longitudinal Clinical Records), a framework that incorporate instruction-response pairs grounding to different parts of a patient's record as a critical dimension in both instruction evaluation and tuning for longitudinal clinical records. We develop TIMER-Bench, the first time-aware benchmark that evaluates temporal reasoning capabilities over longitudinal EHRs, as well as TIMER-Instruct, an instruction-tuning methodology for LLMs to learn reasoning over time. We demonstrate that models fine-tuned with TIMER-Instruct improve performance by 7.3% on human-generated benchmarks and 9.2% on TIMER-Bench, indicating that temporal instruction-tuning improves model performance for reasoning over EHR.
Biomedical Visual Instruction Tuning with Clinician Preference Alignment
Jun 19, 2024



Abstract:Recent advancements in multimodal foundation models have showcased impressive capabilities in understanding and reasoning with visual and textual information. Adapting these foundation models trained for general usage to specialized domains like biomedicine requires large-scale domain-specific instruction datasets. While existing works have explored curating such datasets automatically, the resultant datasets are not explicitly aligned with domain expertise. In this work, we propose a data-centric framework, Biomedical Visual Instruction Tuning with Clinician Preference Alignment (BioMed-VITAL), that incorporates clinician preferences into both stages of generating and selecting instruction data for tuning biomedical multimodal foundation models. First, during the generation stage, we prompt the GPT-4V generator with a diverse set of clinician-selected demonstrations for preference-aligned data candidate generation. Then, during the selection phase, we train a separate selection model, which explicitly distills clinician and policy-guided model preferences into a rating function to select high-quality data for medical instruction tuning. Results show that the model tuned with the instruction-following data from our method demonstrates a significant improvement in open visual chat (18.5% relatively) and medical VQA (win rate up to 81.73%). Our instruction-following data and models are available at BioMed-VITAL.github.io.
TACCO: Task-guided Co-clustering of Clinical Concepts and Patient Visits for Disease Subtyping based on EHR Data
Jun 14, 2024



Abstract:The growing availability of well-organized Electronic Health Records (EHR) data has enabled the development of various machine learning models towards disease risk prediction. However, existing risk prediction methods overlook the heterogeneity of complex diseases, failing to model the potential disease subtypes regarding their corresponding patient visits and clinical concept subgroups. In this work, we introduce TACCO, a novel framework that jointly discovers clusters of clinical concepts and patient visits based on a hypergraph modeling of EHR data. Specifically, we develop a novel self-supervised co-clustering framework that can be guided by the risk prediction task of specific diseases. Furthermore, we enhance the hypergraph model of EHR data with textual embeddings and enforce the alignment between the clusters of clinical concepts and patient visits through a contrastive objective. Comprehensive experiments conducted on the public MIMIC-III dataset and Emory internal CRADLE dataset over the downstream clinical tasks of phenotype classification and cardiovascular risk prediction demonstrate an average 31.25% performance improvement compared to traditional ML baselines and a 5.26% improvement on top of the vanilla hypergraph model without our co-clustering mechanism. In-depth model analysis, clustering results analysis, and clinical case studies further validate the improved utilities and insightful interpretations delivered by TACCO. Code is available at https://github.com/PericlesHat/TACCO.
LLMs-based Few-Shot Disease Predictions using EHR: A Novel Approach Combining Predictive Agent Reasoning and Critical Agent Instruction
Mar 19, 2024Abstract:Electronic health records (EHRs) contain valuable patient data for health-related prediction tasks, such as disease prediction. Traditional approaches rely on supervised learning methods that require large labeled datasets, which can be expensive and challenging to obtain. In this study, we investigate the feasibility of applying Large Language Models (LLMs) to convert structured patient visit data (e.g., diagnoses, labs, prescriptions) into natural language narratives. We evaluate the zero-shot and few-shot performance of LLMs using various EHR-prediction-oriented prompting strategies. Furthermore, we propose a novel approach that utilizes LLM agents with different roles: a predictor agent that makes predictions and generates reasoning processes and a critic agent that analyzes incorrect predictions and provides guidance for improving the reasoning of the predictor agent. Our results demonstrate that with the proposed approach, LLMs can achieve decent few-shot performance compared to traditional supervised learning methods in EHR-based disease predictions, suggesting its potential for health-oriented applications.
Microstructures and Accuracy of Graph Recall by Large Language Models
Feb 20, 2024



Abstract:Graphs data is crucial for many applications, and much of it exists in the relations described in textual format. As a result, being able to accurately recall and encode a graph described in earlier text is a basic yet pivotal ability that LLMs need to demonstrate if they are to perform reasoning tasks that involve graph-structured information. Human performance at graph recall has been studied by cognitive scientists for decades, and has been found to often exhibit certain structural patterns of bias that align with human handling of social relationships. To date, however, we know little about how LLMs behave in analogous graph recall tasks: do their recalled graphs also exhibit certain biased patterns, and if so, how do they compare with humans and affect other graph reasoning tasks? In this work, we perform the first systematical study of graph recall by LLMs, investigating the accuracy and biased microstructures (local structural patterns) in their recall. We find that LLMs not only underperform often in graph recall, but also tend to favor more triangles and alternating 2-paths. Moreover, we find that more advanced LLMs have a striking dependence on the domain that a real-world graph comes from -- by yielding the best recall accuracy when the graph is narrated in a language style consistent with its original domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge