Wei Jin
Department of Computer Science, Emory University, Atlanta, GA, USA
Exposing Vulnerabilities in Explanation for Time Series Classifiers via Dual-Target Attacks
Feb 02, 2026Abstract:Interpretable time series deep learning systems are often assessed by checking temporal consistency on explanations, implicitly treating this as evidence of robustness. We show that this assumption can fail: Predictions and explanations can be adversarially decoupled, enabling targeted misclassification while the explanation remains plausible and consistent with a chosen reference rationale. We propose TSEF (Time Series Explanation Fooler), a dual-target attack that jointly manipulates the classifier and explainer outputs. In contrast to single-objective misclassification attacks that disrupt explanation and spread attribution mass broadly, TSEF achieves targeted prediction changes while keeping explanations consistent with the reference. Across multiple datasets and explainer backbones, our results consistently reveal that explanation stability is a misleading proxy for decision robustness and motivate coupling-aware robustness evaluations for trustworthy time series tasks.
Patient-Conditioned Adaptive Offsets for Reliable Diagnosis across Subgroups
Jan 19, 2026Abstract:AI models for medical diagnosis often exhibit uneven performance across patient populations due to heterogeneity in disease prevalence, imaging appearance, and clinical risk profiles. Existing algorithmic fairness approaches typically seek to reduce such disparities by suppressing sensitive attributes. However, in medical settings these attributes often carry essential diagnostic information, and removing them can degrade accuracy and reliability, particularly in high-stakes applications. In contrast, clinical decision making explicitly incorporates patient context when interpreting diagnostic evidence, suggesting a different design direction for subgroup-aware models. In this paper, we introduce HyperAdapt, a patient-conditioned adaptation framework that improves subgroup reliability while maintaining a shared diagnostic model. Clinically relevant attributes such as age and sex are encoded into a compact embedding and used to condition a hypernetwork-style module, which generates small residual modulation parameters for selected layers of a shared backbone. This design preserves the general medical knowledge learned by the backbone while enabling targeted adjustments that reflect patient-specific variability. To ensure efficiency and robustness, adaptations are constrained through low-rank and bottlenecked parameterizations, limiting both model complexity and computational overhead. Experiments across multiple public medical imaging benchmarks demonstrate that the proposed approach consistently improves subgroup-level performance without sacrificing overall accuracy. On the PAD-UFES-20 dataset, our method outperforms the strongest competing baseline by 4.1% in recall and 4.4% in F1 score, with larger gains observed for underrepresented patient populations.
EpiQAL: Benchmarking Large Language Models in Epidemiological Question Answering for Enhanced Alignment and Reasoning
Jan 06, 2026Abstract:Reliable epidemiological reasoning requires synthesizing study evidence to infer disease burden, transmission dynamics, and intervention effects at the population level. Existing medical question answering benchmarks primarily emphasize clinical knowledge or patient-level reasoning, yet few systematically evaluate evidence-grounded epidemiological inference. We present EpiQAL, the first diagnostic benchmark for epidemiological question answering across diverse diseases, comprising three subsets built from open-access literature. The subsets respectively evaluate text-grounded factual recall, multi-step inference linking document evidence with epidemiological principles, and conclusion reconstruction with the Discussion section withheld. Construction combines expert-designed taxonomy guidance, multi-model verification, and retrieval-based difficulty control. Experiments on ten open models reveal that current LLMs show limited performance on epidemiological reasoning, with multi-step inference posing the greatest challenge. Model rankings shift across subsets, and scale alone does not predict success. Chain-of-Thought prompting benefits multi-step inference but yields mixed results elsewhere. EpiQAL provides fine-grained diagnostic signals for evidence grounding, inferential reasoning, and conclusion reconstruction.
STReasoner: Empowering LLMs for Spatio-Temporal Reasoning in Time Series via Spatial-Aware Reinforcement Learning
Jan 06, 2026Abstract:Spatio-temporal reasoning in time series involves the explicit synthesis of temporal dynamics, spatial dependencies, and textual context. This capability is vital for high-stakes decision-making in systems such as traffic networks, power grids, and disease propagation. However, the field remains underdeveloped because most existing works prioritize predictive accuracy over reasoning. To address the gap, we introduce ST-Bench, a benchmark consisting of four core tasks, including etiological reasoning, entity identification, correlation reasoning, and in-context forecasting, developed via a network SDE-based multi-agent data synthesis pipeline. We then propose STReasoner, which empowers LLM to integrate time series, graph structure, and text for explicit reasoning. To promote spatially grounded logic, we introduce S-GRPO, a reinforcement learning algorithm that rewards performance gains specifically attributable to spatial information. Experiments show that STReasoner achieves average accuracy gains between 17% and 135% at only 0.004X the cost of proprietary models and generalizes robustly to real-world data.
An Adaptive Machine Learning Triage Framework for Predicting Alzheimer's Disease Progression
Nov 10, 2025Abstract:Accurate predictions of conversion from mild cognitive impairment (MCI) to Alzheimer's disease (AD) can enable effective personalized therapy. While cognitive tests and clinical data are routinely collected, they lack the predictive power of PET scans and CSF biomarker analysis, which are prohibitively expensive to obtain for every patient. To address this cost-accuracy dilemma, we design a two-stage machine learning framework that selectively obtains advanced, costly features based on their predicted "value of information". We apply our framework to predict AD progression for MCI patients using data from the Alzheimer's Disease Neuroimaging Initiative (ADNI). Our framework reduces the need for advanced testing by 20% while achieving a test AUROC of 0.929, comparable to the model that uses both basic and advanced features (AUROC=0.915, p=0.1010). We also provide an example interpretability analysis showing how one may explain the triage decision. Our work presents an interpretable, data-driven framework that optimizes AD diagnostic pathways and balances accuracy with cost, representing a step towards making early, reliable AD prediction more accessible in real-world practice. Future work should consider multiple categories of advanced features and larger-scale validation.
Generalist vs Specialist Time Series Foundation Models: Investigating Potential Emergent Behaviors in Assessing Human Health Using PPG Signals
Oct 16, 2025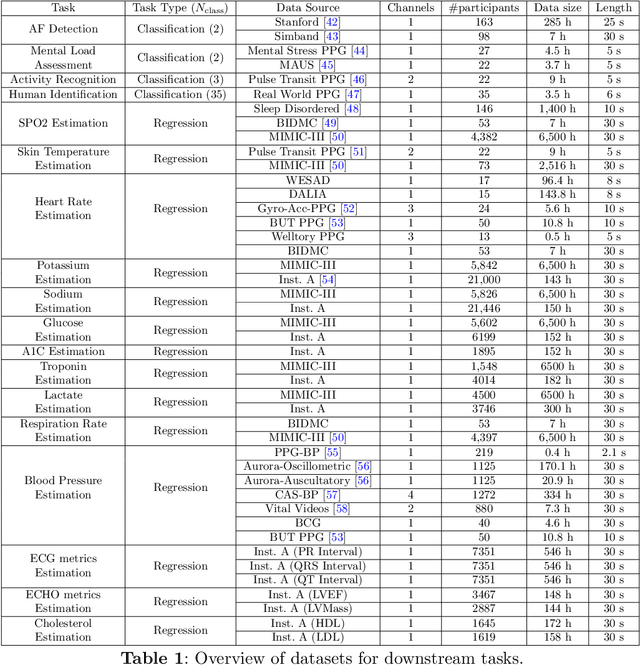
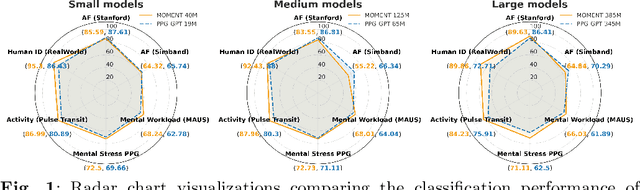
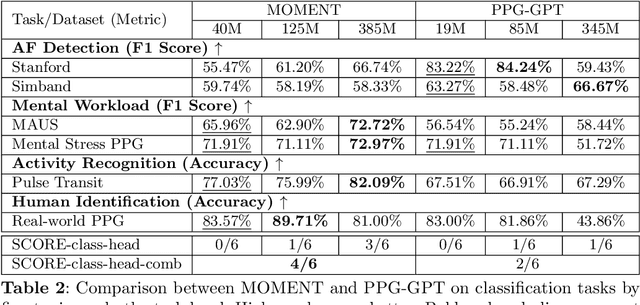
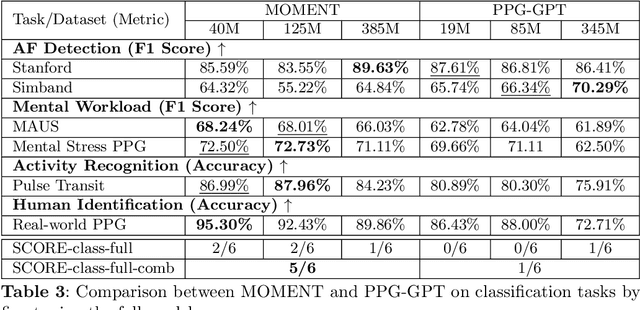
Abstract:Foundation models are large-scale machine learning models that are pre-trained on massive amounts of data and can be adapted for various downstream tasks. They have been extensively applied to tasks in Natural Language Processing and Computer Vision with models such as GPT, BERT, and CLIP. They are now also increasingly gaining attention in time-series analysis, particularly for physiological sensing. However, most time series foundation models are specialist models - with data in pre-training and testing of the same type, such as Electrocardiogram, Electroencephalogram, and Photoplethysmogram (PPG). Recent works, such as MOMENT, train a generalist time series foundation model with data from multiple domains, such as weather, traffic, and electricity. This paper aims to conduct a comprehensive benchmarking study to compare the performance of generalist and specialist models, with a focus on PPG signals. Through an extensive suite of total 51 tasks covering cardiac state assessment, laboratory value estimation, and cross-modal inference, we comprehensively evaluate both models across seven dimensions, including win score, average performance, feature quality, tuning gain, performance variance, transferability, and scalability. These metrics jointly capture not only the models' capability but also their adaptability, robustness, and efficiency under different fine-tuning strategies, providing a holistic understanding of their strengths and limitations for diverse downstream scenarios. In a full-tuning scenario, we demonstrate that the specialist model achieves a 27% higher win score. Finally, we provide further analysis on generalization, fairness, attention visualizations, and the importance of training data choice.
Can Large Language Models Adequately Perform Symbolic Reasoning Over Time Series?
Aug 05, 2025Abstract:Uncovering hidden symbolic laws from time series data, as an aspiration dating back to Kepler's discovery of planetary motion, remains a core challenge in scientific discovery and artificial intelligence. While Large Language Models show promise in structured reasoning tasks, their ability to infer interpretable, context-aligned symbolic structures from time series data is still underexplored. To systematically evaluate this capability, we introduce SymbolBench, a comprehensive benchmark designed to assess symbolic reasoning over real-world time series across three tasks: multivariate symbolic regression, Boolean network inference, and causal discovery. Unlike prior efforts limited to simple algebraic equations, SymbolBench spans a diverse set of symbolic forms with varying complexity. We further propose a unified framework that integrates LLMs with genetic programming to form a closed-loop symbolic reasoning system, where LLMs act both as predictors and evaluators. Our empirical results reveal key strengths and limitations of current models, highlighting the importance of combining domain knowledge, context alignment, and reasoning structure to improve LLMs in automated scientific discovery.
TimeRecipe: A Time-Series Forecasting Recipe via Benchmarking Module Level Effectiveness
Jun 06, 2025Abstract:Time-series forecasting is an essential task with wide real-world applications across domains. While recent advances in deep learning have enabled time-series forecasting models with accurate predictions, there remains considerable debate over which architectures and design components, such as series decomposition or normalization, are most effective under varying conditions. Existing benchmarks primarily evaluate models at a high level, offering limited insight into why certain designs work better. To mitigate this gap, we propose TimeRecipe, a unified benchmarking framework that systematically evaluates time-series forecasting methods at the module level. TimeRecipe conducts over 10,000 experiments to assess the effectiveness of individual components across a diverse range of datasets, forecasting horizons, and task settings. Our results reveal that exhaustive exploration of the design space can yield models that outperform existing state-of-the-art methods and uncover meaningful intuitions linking specific design choices to forecasting scenarios. Furthermore, we release a practical toolkit within TimeRecipe that recommends suitable model architectures based on these empirical insights. The benchmark is available at: https://github.com/AdityaLab/TimeRecipe.
SeizureFormer: A Transformer Model for IEA-Based Seizure Risk Forecasting
Apr 24, 2025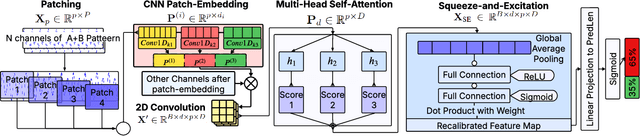
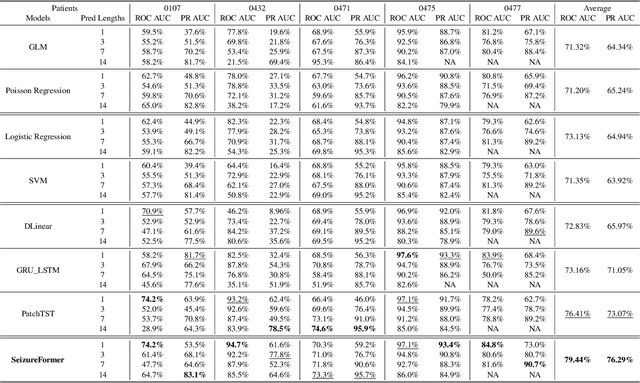
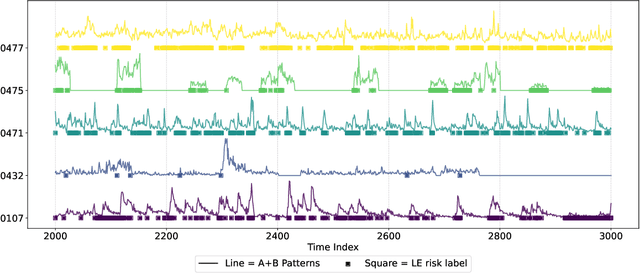
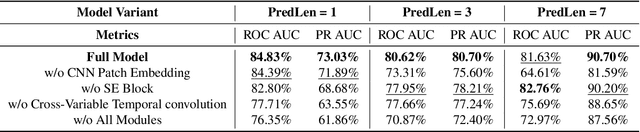
Abstract:We present SeizureFormer, a Transformer-based model for long-term seizure risk forecasting using interictal epileptiform activity (IEA) surrogate biomarkers and long episode (LE) biomarkers from responsive neurostimulation (RNS) systems. Unlike raw scalp EEG-based models, SeizureFormer leverages structured, clinically relevant features and integrates CNN-based patch embedding, multi-head self-attention, and squeeze-and-excitation blocks to model both short-term dynamics and long-term seizure cycles. Tested across five patients and multiple prediction windows (1 to 14 days), SeizureFormer achieved state-of-the-art performance with mean ROC AUC of 79.44 percent and mean PR AUC of 76.29 percent. Compared to statistical, machine learning, and deep learning baselines, it demonstrates enhanced generalizability and seizure risk forecasting performance under class imbalance. This work supports future clinical integration of interpretable and robust seizure forecasting tools for personalized epilepsy management.
Graph ODEs and Beyond: A Comprehensive Survey on Integrating Differential Equations with Graph Neural Networks
Mar 29, 2025Abstract:Graph Neural Networks (GNNs) and differential equations (DEs) are two rapidly advancing areas of research that have shown remarkable synergy in recent years. GNNs have emerged as powerful tools for learning on graph-structured data, while differential equations provide a principled framework for modeling continuous dynamics across time and space. The intersection of these fields has led to innovative approaches that leverage the strengths of both, enabling applications in physics-informed learning, spatiotemporal modeling, and scientific computing. This survey aims to provide a comprehensive overview of the burgeoning research at the intersection of GNNs and DEs. We will categorize existing methods, discuss their underlying principles, and highlight their applications across domains such as molecular modeling, traffic prediction, and epidemic spreading. Furthermore, we identify open challenges and outline future research directions to advance this interdisciplinary field. A comprehensive paper list is provided at https://github.com/Emory-Melody/Awesome-Graph-NDEs. This survey serves as a resource for researchers and practitioners seeking to understand and contribute to the fusion of GNNs and DEs
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge