Wenzhuo Tang
Sub-graph Based Diffusion Model for Link Prediction
Sep 13, 2024Abstract:Denoising Diffusion Probabilistic Models (DDPMs) represent a contemporary class of generative models with exceptional qualities in both synthesis and maximizing the data likelihood. These models work by traversing a forward Markov Chain where data is perturbed, followed by a reverse process where a neural network learns to undo the perturbations and recover the original data. There have been increasing efforts exploring the applications of DDPMs in the graph domain. However, most of them have focused on the generative perspective. In this paper, we aim to build a novel generative model for link prediction. In particular, we treat link prediction between a pair of nodes as a conditional likelihood estimation of its enclosing sub-graph. With a dedicated design to decompose the likelihood estimation process via the Bayesian formula, we are able to separate the estimation of sub-graph structure and its node features. Such designs allow our model to simultaneously enjoy the advantages of inductive learning and the strong generalization capability. Remarkably, comprehensive experiments across various datasets validate that our proposed method presents numerous advantages: (1) transferability across datasets without retraining, (2) promising generalization on limited training data, and (3) robustness against graph adversarial attacks.
Cross-Domain Graph Data Scaling: A Showcase with Diffusion Models
Jun 04, 2024Abstract:Models for natural language and images benefit from data scaling behavior: the more data fed into the model, the better they perform. This 'better with more' phenomenon enables the effectiveness of large-scale pre-training on vast amounts of data. However, current graph pre-training methods struggle to scale up data due to heterogeneity across graphs. To achieve effective data scaling, we aim to develop a general model that is able to capture diverse data patterns of graphs and can be utilized to adaptively help the downstream tasks. To this end, we propose UniAug, a universal graph structure augmentor built on a diffusion model. We first pre-train a discrete diffusion model on thousands of graphs across domains to learn the graph structural patterns. In the downstream phase, we provide adaptive enhancement by conducting graph structure augmentation with the help of the pre-trained diffusion model via guided generation. By leveraging the pre-trained diffusion model for structure augmentation, we consistently achieve performance improvements across various downstream tasks in a plug-and-play manner. To the best of our knowledge, this study represents the first demonstration of a data-scaling graph structure augmentor on graphs across domains.
Graph Machine Learning in the Era of Large Language Models
Apr 23, 2024Abstract:Graphs play an important role in representing complex relationships in various domains like social networks, knowledge graphs, and molecular discovery. With the advent of deep learning, Graph Neural Networks (GNNs) have emerged as a cornerstone in Graph Machine Learning (Graph ML), facilitating the representation and processing of graph structures. Recently, LLMs have demonstrated unprecedented capabilities in language tasks and are widely adopted in a variety of applications such as computer vision and recommender systems. This remarkable success has also attracted interest in applying LLMs to the graph domain. Increasing efforts have been made to explore the potential of LLMs in advancing Graph ML's generalization, transferability, and few-shot learning ability. Meanwhile, graphs, especially knowledge graphs, are rich in reliable factual knowledge, which can be utilized to enhance the reasoning capabilities of LLMs and potentially alleviate their limitations such as hallucinations and the lack of explainability. Given the rapid progress of this research direction, a systematic review summarizing the latest advancements for Graph ML in the era of LLMs is necessary to provide an in-depth understanding to researchers and practitioners. Therefore, in this survey, we first review the recent developments in Graph ML. We then explore how LLMs can be utilized to enhance the quality of graph features, alleviate the reliance on labeled data, and address challenges such as graph heterogeneity and out-of-distribution (OOD) generalization. Afterward, we delve into how graphs can enhance LLMs, highlighting their abilities to enhance LLM pre-training and inference. Furthermore, we investigate various applications and discuss the potential future directions in this promising field.
Graph Foundation Models
Feb 06, 2024



Abstract:Graph Foundation Model (GFM) is a new trending research topic in the graph domain, aiming to develop a graph model capable of generalizing across different graphs and tasks. However, a versatile GFM has not yet been achieved. The key challenge in building GFM is how to enable positive transfer across graphs with diverse structural patterns. Inspired by the existing foundation models in the CV and NLP domains, we propose a novel perspective for the GFM development by advocating for a "graph vocabulary", in which the basic transferable units underlying graphs encode the invariance on graphs. We ground the graph vocabulary construction from essential aspects including network analysis, theoretical foundations, and stability. Such a vocabulary perspective can potentially advance the future GFM design following the neural scaling laws.
Single-Cell Multimodal Prediction via Transformers
Mar 01, 2023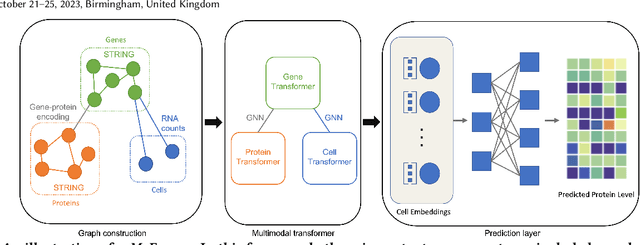
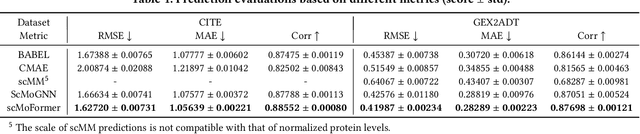
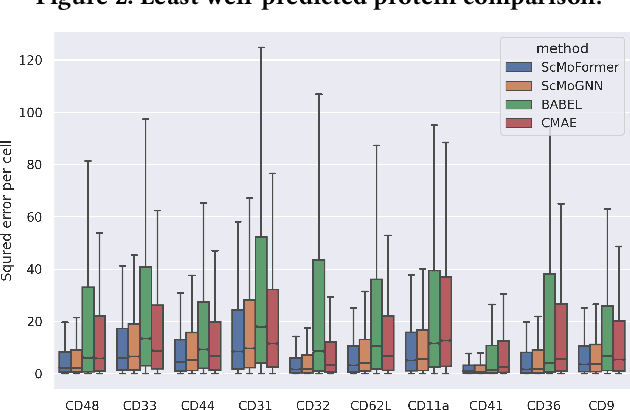
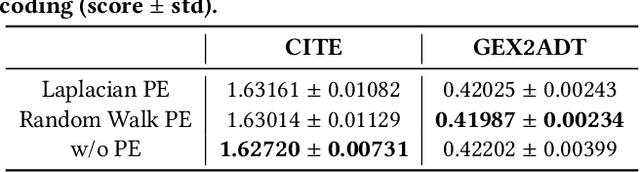
Abstract:The recent development of multimodal single-cell technology has made the possibility of acquiring multiple omics data from individual cells, thereby enabling a deeper understanding of cellular states and dynamics. Nevertheless, the proliferation of multimodal single-cell data also introduces tremendous challenges in modeling the complex interactions among different modalities. The recently advanced methods focus on constructing static interaction graphs and applying graph neural networks (GNNs) to learn from multimodal data. However, such static graphs can be suboptimal as they do not take advantage of the downstream task information; meanwhile GNNs also have some inherent limitations when deeply stacking GNN layers. To tackle these issues, in this work, we investigate how to leverage transformers for multimodal single-cell data in an end-to-end manner while exploiting downstream task information. In particular, we propose a scMoFormer framework which can readily incorporate external domain knowledge and model the interactions within each modality and cross modalities. Extensive experiments demonstrate that scMoFormer achieves superior performance on various benchmark datasets. Note that scMoFormer won a Kaggle silver medal with the rank of $24\ /\ 1221$ (Top 2%) without ensemble in a NeurIPS 2022 competition. Our implementation is publicly available at Github.
Single Cells Are Spatial Tokens: Transformers for Spatial Transcriptomic Data Imputation
Feb 06, 2023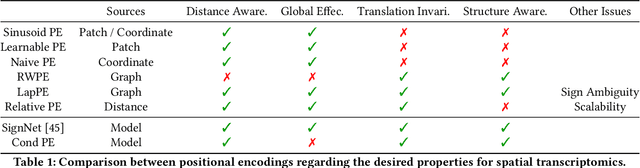
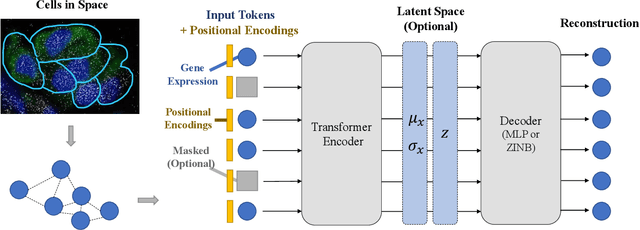
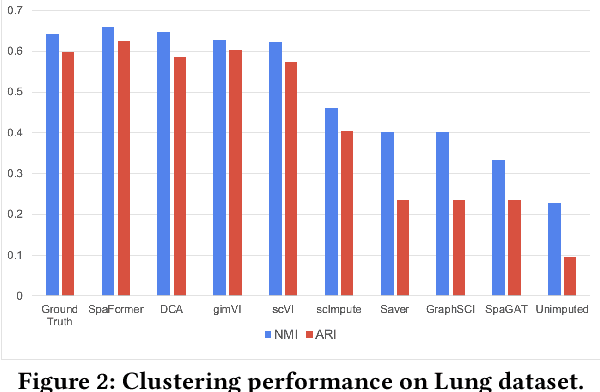
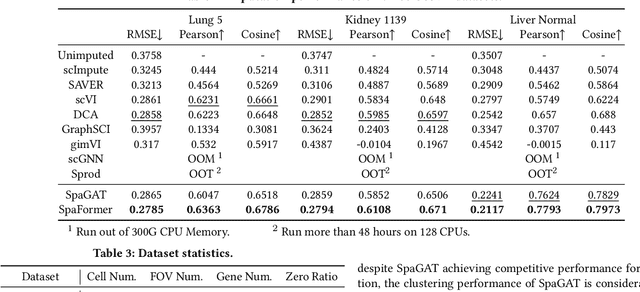
Abstract:Spatially resolved transcriptomics brings exciting breakthroughs to single-cell analysis by providing physical locations along with gene expression. However, as a cost of the extremely high spatial resolution, the cellular level spatial transcriptomic data suffer significantly from missing values. While a standard solution is to perform imputation on the missing values, most existing methods either overlook spatial information or only incorporate localized spatial context without the ability to capture long-range spatial information. Using multi-head self-attention mechanisms and positional encoding, transformer models can readily grasp the relationship between tokens and encode location information. In this paper, by treating single cells as spatial tokens, we study how to leverage transformers to facilitate spatial tanscriptomics imputation. In particular, investigate the following two key questions: (1) $\textit{how to encode spatial information of cells in transformers}$, and (2) $\textit{ how to train a transformer for transcriptomic imputation}$. By answering these two questions, we present a transformer-based imputation framework, SpaFormer, for cellular-level spatial transcriptomic data. Extensive experiments demonstrate that SpaFormer outperforms existing state-of-the-art imputation algorithms on three large-scale datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge