Feng Shi
KernelEvolve: Scaling Agentic Kernel Coding for Heterogeneous AI Accelerators at Meta
Dec 30, 2025Abstract:Making deep learning recommendation model (DLRM) training and inference fast and efficient is important. However, this presents three key system challenges - model architecture diversity, kernel primitive diversity, and hardware generation and architecture heterogeneity. This paper presents KernelEvolve-an agentic kernel coding framework-to tackle heterogeneity at-scale for DLRM. KernelEvolve is designed to take kernel specifications as input and automate the process of kernel generation and optimization for recommendation model across heterogeneous hardware architectures. KernelEvolve does so by operating at multiple programming abstractions, from Triton and CuTe DSL to low-level hardware agnostic languages, spanning the full hardware-software optimization stack. The kernel optimization process is described as graph-based search with selection policy, universal operator, fitness function, and termination rule, dynamically adapts to runtime execution context through retrieval-augmented prompt synthesis. We designed, implemented, and deployed KernelEvolve to optimize a wide variety of production recommendation models across generations of NVIDIA and AMD GPUs, as well as Meta's AI accelerators. We validate KernelEvolve on the publicly-available KernelBench suite, achieving 100% pass rate on all 250 problems across three difficulty levels, and 160 PyTorch ATen operators across three heterogeneous hardware platforms, demonstrating 100% correctness. KernelEvolve reduces development time from weeks to hours and achieves substantial performance improvements over PyTorch baselines across diverse production use cases and for heterogeneous AI systems at-scale. Beyond performance efficiency improvements, KernelEvolve significantly mitigates the programmability barrier for new AI hardware by enabling automated kernel generation for in-house developed AI hardware.
AttentionDrag: Exploiting Latent Correlation Knowledge in Pre-trained Diffusion Models for Image Editing
Jun 16, 2025



Abstract:Traditional point-based image editing methods rely on iterative latent optimization or geometric transformations, which are either inefficient in their processing or fail to capture the semantic relationships within the image. These methods often overlook the powerful yet underutilized image editing capabilities inherent in pre-trained diffusion models. In this work, we propose a novel one-step point-based image editing method, named AttentionDrag, which leverages the inherent latent knowledge and feature correlations within pre-trained diffusion models for image editing tasks. This framework enables semantic consistency and high-quality manipulation without the need for extensive re-optimization or retraining. Specifically, we reutilize the latent correlations knowledge learned by the self-attention mechanism in the U-Net module during the DDIM inversion process to automatically identify and adjust relevant image regions, ensuring semantic validity and consistency. Additionally, AttentionDrag adaptively generates masks to guide the editing process, enabling precise and context-aware modifications with friendly interaction. Our results demonstrate a performance that surpasses most state-of-the-art methods with significantly faster speeds, showing a more efficient and semantically coherent solution for point-based image editing tasks.
A 2D Semantic-Aware Position Encoding for Vision Transformers
May 14, 2025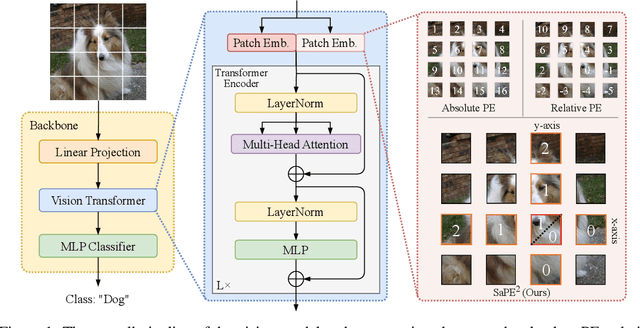
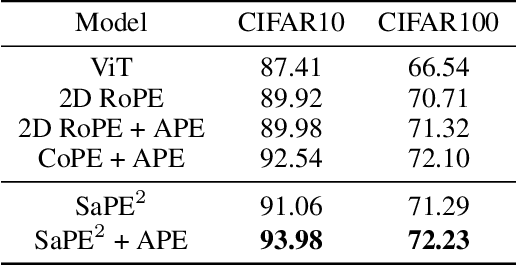
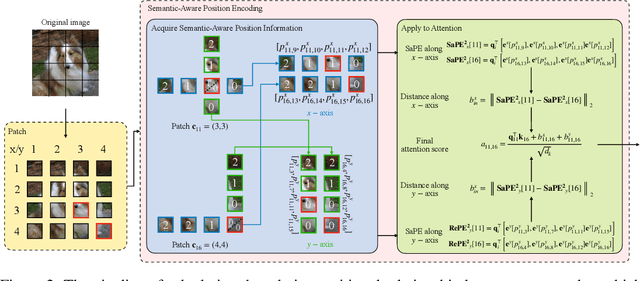
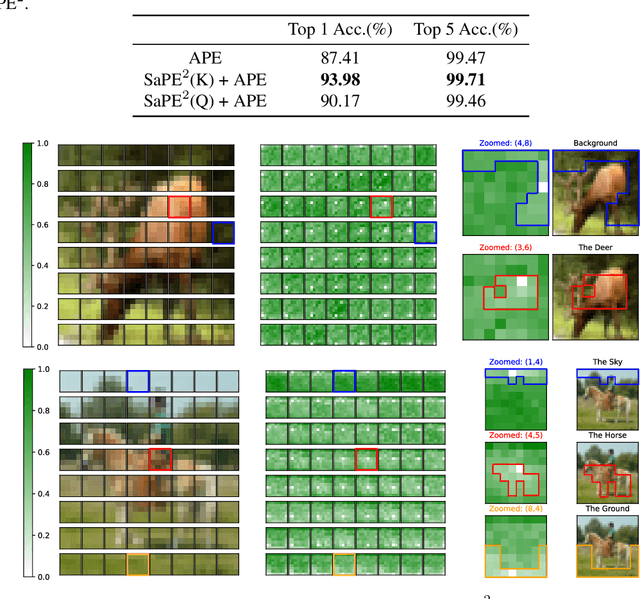
Abstract:Vision transformers have demonstrated significant advantages in computer vision tasks due to their ability to capture long-range dependencies and contextual relationships through self-attention. However, existing position encoding techniques, which are largely borrowed from natural language processing, fail to effectively capture semantic-aware positional relationships between image patches. Traditional approaches like absolute position encoding and relative position encoding primarily focus on 1D linear position relationship, often neglecting the semantic similarity between distant yet contextually related patches. These limitations hinder model generalization, translation equivariance, and the ability to effectively handle repetitive or structured patterns in images. In this paper, we propose 2-Dimensional Semantic-Aware Position Encoding ($\text{SaPE}^2$), a novel position encoding method with semantic awareness that dynamically adapts position representations by leveraging local content instead of fixed linear position relationship or spatial coordinates. Our method enhances the model's ability to generalize across varying image resolutions and scales, improves translation equivariance, and better aggregates features for visually similar but spatially distant patches. By integrating $\text{SaPE}^2$ into vision transformers, we bridge the gap between position encoding and perceptual similarity, thereby improving performance on computer vision tasks.
ChatZero:Zero-shot Cross-Lingual Dialogue Generation via Pseudo-Target Language
Aug 16, 2024Abstract:Although large language models(LLMs) show amazing capabilities, among various exciting applications discovered for LLMs fall short in other low-resource languages. Besides, most existing methods depend on large-scale dialogue corpora and thus building systems for dialogue generation in a zero-shot scenario remains a considerable challenge. To address this challenge, we propose a novel end-to-end zero-shot dialogue generation model ChatZero based on cross-lingual code-switching method. First, we construct code-switching language and pseudo-target language with placeholders. Then for cross-lingual semantic transfer, we employ unsupervised contrastive learning to minimize the semantics gap of the source language, code-switching language, and pseudo-target language that are mutually positive examples in the high dimensional semantic space. Experiments on the multilingual DailyDialog and DSTC7-AVSD datasets demonstrate that ChatZero can achieve more than 90\% of the original performance under the zero-shot case compared to supervised learning, and achieve state-of-the-art performance compared with other baselines.
* ECAI2024
LipidBERT: A Lipid Language Model Pre-trained on METiS de novo Lipid Library
Aug 12, 2024



Abstract:In this study, we generate and maintain a database of 10 million virtual lipids through METiS's in-house de novo lipid generation algorithms and lipid virtual screening techniques. These virtual lipids serve as a corpus for pre-training, lipid representation learning, and downstream task knowledge transfer, culminating in state-of-the-art LNP property prediction performance. We propose LipidBERT, a BERT-like model pre-trained with the Masked Language Model (MLM) and various secondary tasks. Additionally, we compare the performance of embeddings generated by LipidBERT and PhatGPT, our GPT-like lipid generation model, on downstream tasks. The proposed bilingual LipidBERT model operates in two languages: the language of ionizable lipid pre-training, using in-house dry-lab lipid structures, and the language of LNP fine-tuning, utilizing in-house LNP wet-lab data. This dual capability positions LipidBERT as a key AI-based filter for future screening tasks, including new versions of METiS de novo lipid libraries and, more importantly, candidates for in vivo testing for orgran-targeting LNPs. To the best of our knowledge, this is the first successful demonstration of the capability of a pre-trained language model on virtual lipids and its effectiveness in downstream tasks using web-lab data. This work showcases the clever utilization of METiS's in-house de novo lipid library as well as the power of dry-wet lab integration.
Knowledge-Guided Prompt Learning for Lifespan Brain MR Image Segmentation
Jul 31, 2024



Abstract:Automatic and accurate segmentation of brain MR images throughout the human lifespan into tissue and structure is crucial for understanding brain development and diagnosing diseases. However, challenges arise from the intricate variations in brain appearance due to rapid early brain development, aging, and disorders, compounded by the limited availability of manually-labeled datasets. In response, we present a two-step segmentation framework employing Knowledge-Guided Prompt Learning (KGPL) for brain MRI. Specifically, we first pre-train segmentation models on large-scale datasets with sub-optimal labels, followed by the incorporation of knowledge-driven embeddings learned from image-text alignment into the models. The introduction of knowledge-wise prompts captures semantic relationships between anatomical variability and biological processes, enabling models to learn structural feature embeddings across diverse age groups. Experimental findings demonstrate the superiority and robustness of our proposed method, particularly noticeable when employing Swin UNETR as the backbone. Our approach achieves average DSC values of 95.17% and 94.19% for brain tissue and structure segmentation, respectively. Our code is available at https://github.com/TL9792/KGPL.
Hunting imaging biomarkers in pulmonary fibrosis: Benchmarks of the AIIB23 challenge
Dec 21, 2023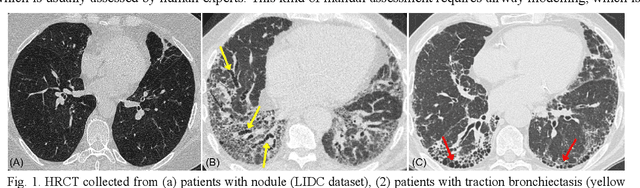
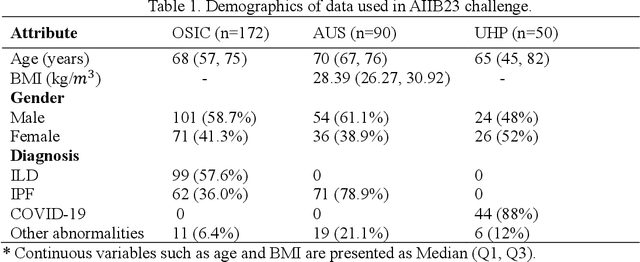
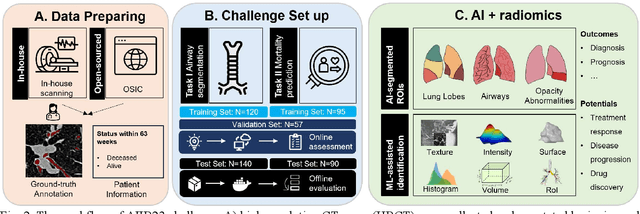
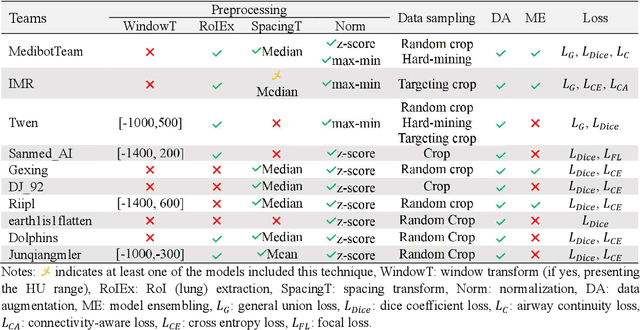
Abstract:Airway-related quantitative imaging biomarkers are crucial for examination, diagnosis, and prognosis in pulmonary diseases. However, the manual delineation of airway trees remains prohibitively time-consuming. While significant efforts have been made towards enhancing airway modelling, current public-available datasets concentrate on lung diseases with moderate morphological variations. The intricate honeycombing patterns present in the lung tissues of fibrotic lung disease patients exacerbate the challenges, often leading to various prediction errors. To address this issue, the 'Airway-Informed Quantitative CT Imaging Biomarker for Fibrotic Lung Disease 2023' (AIIB23) competition was organized in conjunction with the official 2023 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). The airway structures were meticulously annotated by three experienced radiologists. Competitors were encouraged to develop automatic airway segmentation models with high robustness and generalization abilities, followed by exploring the most correlated QIB of mortality prediction. A training set of 120 high-resolution computerised tomography (HRCT) scans were publicly released with expert annotations and mortality status. The online validation set incorporated 52 HRCT scans from patients with fibrotic lung disease and the offline test set included 140 cases from fibrosis and COVID-19 patients. The results have shown that the capacity of extracting airway trees from patients with fibrotic lung disease could be enhanced by introducing voxel-wise weighted general union loss and continuity loss. In addition to the competitive image biomarkers for prognosis, a strong airway-derived biomarker (Hazard ratio>1.5, p<0.0001) was revealed for survival prognostication compared with existing clinical measurements, clinician assessment and AI-based biomarkers.
Optimizing Chance-Constrained Submodular Problems with Variable Uncertainties
Sep 23, 2023Abstract:Chance constraints are frequently used to limit the probability of constraint violations in real-world optimization problems where the constraints involve stochastic components. We study chance-constrained submodular optimization problems, which capture a wide range of optimization problems with stochastic constraints. Previous studies considered submodular problems with stochastic knapsack constraints in the case where uncertainties are the same for each item that can be selected. However, uncertainty levels are usually variable with respect to the different stochastic components in real-world scenarios, and rigorous analysis for this setting is missing in the context of submodular optimization. This paper provides the first such analysis for this case, where the weights of items have the same expectation but different dispersion. We present greedy algorithms that can obtain a high-quality solution, i.e., a constant approximation ratio to the given optimal solution from the deterministic setting. In the experiments, we demonstrate that the algorithms perform effectively on several chance-constrained instances of the maximum coverage problem and the influence maximization problem.
Fragment and Integrate Network (FIN): A Novel Spatial-Temporal Modeling Based on Long Sequential Behavior for Online Food Ordering Click-Through Rate Prediction
Aug 30, 2023



Abstract:Spatial-temporal information has been proven to be of great significance for click-through rate prediction tasks in online Location-Based Services (LBS), especially in mainstream food ordering platforms such as DoorDash, Uber Eats, Meituan, and Ele.me. Modeling user spatial-temporal preferences with sequential behavior data has become a hot topic in recommendation systems and online advertising. However, most of existing methods either lack the representation of rich spatial-temporal information or only handle user behaviors with limited length, e.g. 100. In this paper, we tackle these problems by designing a new spatial-temporal modeling paradigm named Fragment and Integrate Network (FIN). FIN consists of two networks: (i) Fragment Network (FN) extracts Multiple Sub-Sequences (MSS) from lifelong sequential behavior data, and captures the specific spatial-temporal representation by modeling each MSS respectively. Here both a simplified attention and a complicated attention are adopted to balance the performance gain and resource consumption. (ii) Integrate Network (IN) builds a new integrated sequence by utilizing spatial-temporal interaction on MSS and captures the comprehensive spatial-temporal representation by modeling the integrated sequence with a complicated attention. Both public datasets and production datasets have demonstrated the accuracy and scalability of FIN. Since 2022, FIN has been fully deployed in the recommendation advertising system of Ele.me, one of the most popular online food ordering platforms in China, obtaining 5.7% improvement on Click-Through Rate (CTR) and 7.3% increase on Revenue Per Mille (RPM).
Towards Label Position Bias in Graph Neural Networks
May 25, 2023Abstract:Graph Neural Networks (GNNs) have emerged as a powerful tool for semi-supervised node classification tasks. However, recent studies have revealed various biases in GNNs stemming from both node features and graph topology. In this work, we uncover a new bias - label position bias, which indicates that the node closer to the labeled nodes tends to perform better. We introduce a new metric, the Label Proximity Score, to quantify this bias, and find that it is closely related to performance disparities. To address the label position bias, we propose a novel optimization framework for learning a label position unbiased graph structure, which can be applied to existing GNNs. Extensive experiments demonstrate that our proposed method not only outperforms backbone methods but also significantly mitigates the issue of label position bias in GNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge