Yang Nan
Embedding Perturbation may Better Reflect the Uncertainty in LLM Reasoning
Feb 02, 2026Abstract:Large language Models (LLMs) have achieved significant breakthroughs across diverse domains; however, they can still produce unreliable or misleading outputs. For responsible LLM application, Uncertainty Quantification (UQ) techniques are used to estimate a model's uncertainty about its outputs, indicating the likelihood that those outputs may be problematic. For LLM reasoning tasks, it is essential to estimate the uncertainty not only for the final answer, but also for the intermediate steps of the reasoning, as this can enable more fine-grained and targeted interventions. In this study, we explore what UQ metrics better reflect the LLM's ``intermediate uncertainty''during reasoning. Our study reveals that an LLMs' incorrect reasoning steps tend to contain tokens which are highly sensitive to the perturbations on the preceding token embeddings. In this way, incorrect (uncertain) intermediate steps can be readily identified using this sensitivity score as guidance in practice. In our experiments, we show such perturbation-based metric achieves stronger uncertainty quantification performance compared with baseline methods such as token (generation) probability and token entropy. Besides, different from approaches that rely on multiple sampling, the perturbation-based metrics offer better simplicity and efficiency.
Interpretable Probability Estimation with LLMs via Shapley Reconstruction
Jan 14, 2026Abstract:Large Language Models (LLMs) demonstrate potential to estimate the probability of uncertain events, by leveraging their extensive knowledge and reasoning capabilities. This ability can be applied to support intelligent decision-making across diverse fields, such as financial forecasting and preventive healthcare. However, directly prompting LLMs for probability estimation faces significant challenges: their outputs are often noisy, and the underlying predicting process is opaque. In this paper, we propose PRISM: Probability Reconstruction via Shapley Measures, a framework that brings transparency and precision to LLM-based probability estimation. PRISM decomposes an LLM's prediction by quantifying the marginal contribution of each input factor using Shapley values. These factor-level contributions are then aggregated to reconstruct a calibrated final estimate. In our experiments, we demonstrate PRISM improves predictive accuracy over direct prompting and other baselines, across multiple domains including finance, healthcare, and agriculture. Beyond performance, PRISM provides a transparent prediction pipeline: our case studies visualize how individual factors shape the final estimate, helping build trust in LLM-based decision support systems.
AlphaGo Moment for Model Architecture Discovery
Jul 24, 2025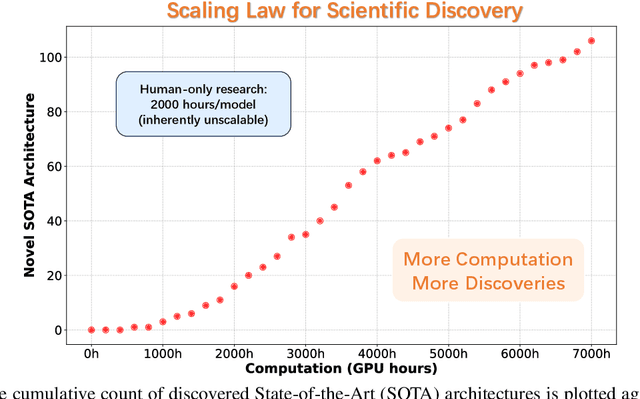
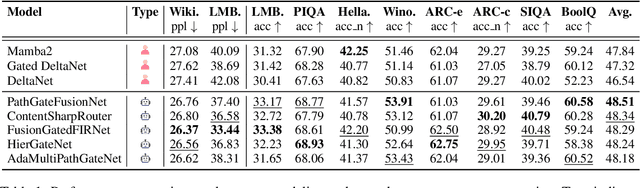
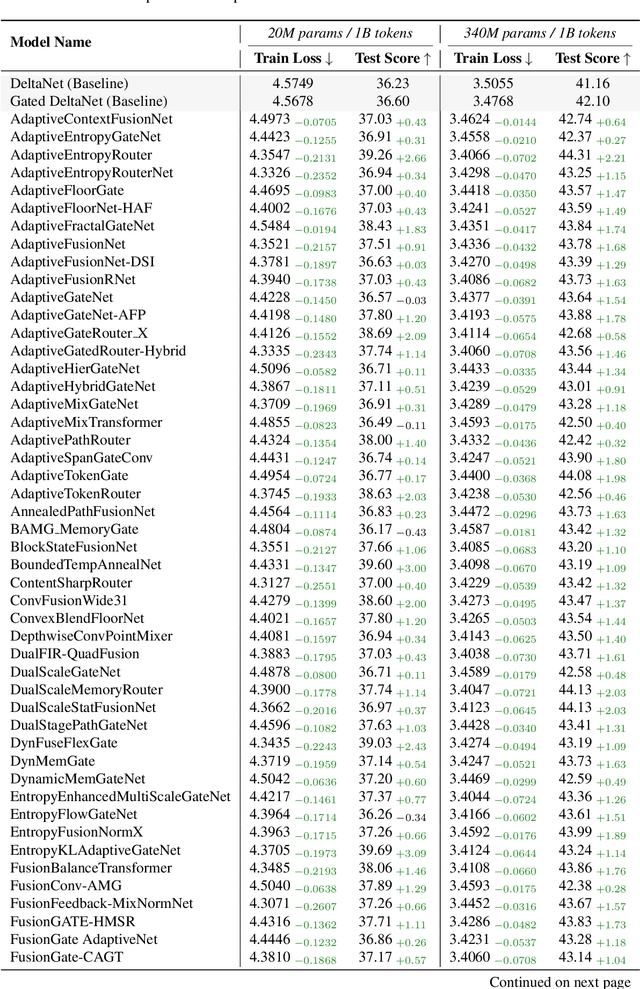
Abstract:While AI systems demonstrate exponentially improving capabilities, the pace of AI research itself remains linearly bounded by human cognitive capacity, creating an increasingly severe development bottleneck. We present ASI-Arch, the first demonstration of Artificial Superintelligence for AI research (ASI4AI) in the critical domain of neural architecture discovery--a fully autonomous system that shatters this fundamental constraint by enabling AI to conduct its own architectural innovation. Moving beyond traditional Neural Architecture Search (NAS), which is fundamentally limited to exploring human-defined spaces, we introduce a paradigm shift from automated optimization to automated innovation. ASI-Arch can conduct end-to-end scientific research in the domain of architecture discovery, autonomously hypothesizing novel architectural concepts, implementing them as executable code, training and empirically validating their performance through rigorous experimentation and past experience. ASI-Arch conducted 1,773 autonomous experiments over 20,000 GPU hours, culminating in the discovery of 106 innovative, state-of-the-art (SOTA) linear attention architectures. Like AlphaGo's Move 37 that revealed unexpected strategic insights invisible to human players, our AI-discovered architectures demonstrate emergent design principles that systematically surpass human-designed baselines and illuminate previously unknown pathways for architectural innovation. Crucially, we establish the first empirical scaling law for scientific discovery itself--demonstrating that architectural breakthroughs can be scaled computationally, transforming research progress from a human-limited to a computation-scalable process. We provide comprehensive analysis of the emergent design patterns and autonomous research capabilities that enabled these breakthroughs, establishing a blueprint for self-accelerating AI systems.
Revisiting Medical Image Retrieval via Knowledge Consolidation
Mar 12, 2025Abstract:As artificial intelligence and digital medicine increasingly permeate healthcare systems, robust governance frameworks are essential to ensure ethical, secure, and effective implementation. In this context, medical image retrieval becomes a critical component of clinical data management, playing a vital role in decision-making and safeguarding patient information. Existing methods usually learn hash functions using bottleneck features, which fail to produce representative hash codes from blended embeddings. Although contrastive hashing has shown superior performance, current approaches often treat image retrieval as a classification task, using category labels to create positive/negative pairs. Moreover, many methods fail to address the out-of-distribution (OOD) issue when models encounter external OOD queries or adversarial attacks. In this work, we propose a novel method to consolidate knowledge of hierarchical features and optimisation functions. We formulate the knowledge consolidation by introducing Depth-aware Representation Fusion (DaRF) and Structure-aware Contrastive Hashing (SCH). DaRF adaptively integrates shallow and deep representations into blended features, and SCH incorporates image fingerprints to enhance the adaptability of positive/negative pairings. These blended features further facilitate OOD detection and content-based recommendation, contributing to a secure AI-driven healthcare environment. Moreover, we present a content-guided ranking to improve the robustness and reproducibility of retrieval results. Our comprehensive assessments demonstrate that the proposed method could effectively recognise OOD samples and significantly outperform existing approaches in medical image retrieval (p<0.05). In particular, our method achieves a 5.6-38.9% improvement in mean Average Precision on the anatomical radiology dataset.
Measuring Data Diversity for Instruction Tuning: A Systematic Analysis and A Reliable Metric
Feb 25, 2025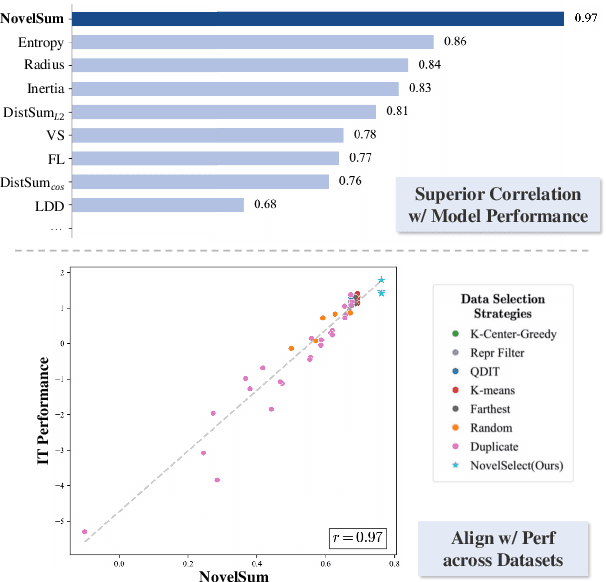
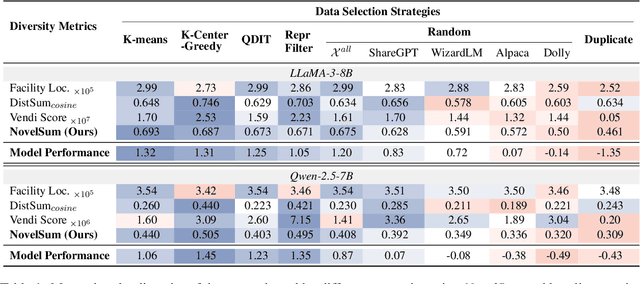
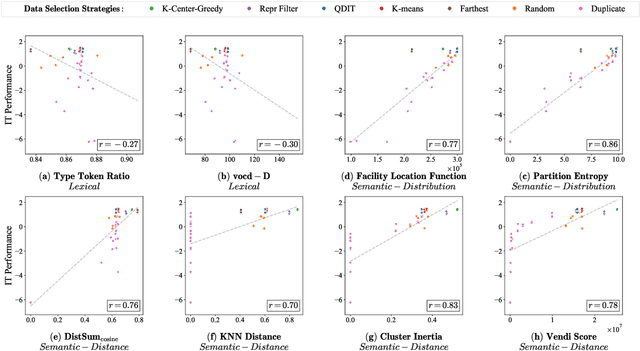
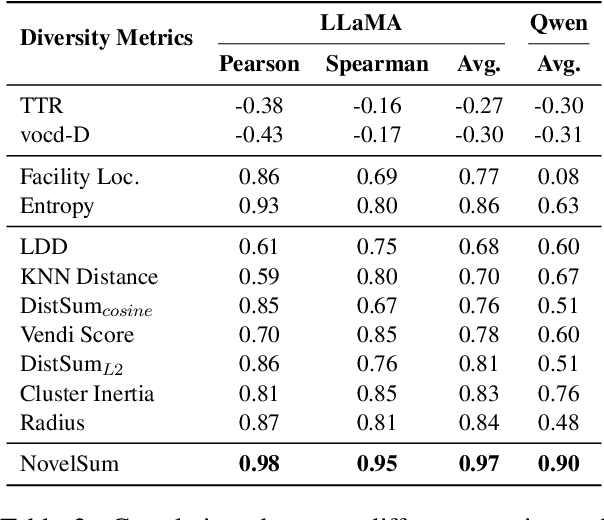
Abstract:Data diversity is crucial for the instruction tuning of large language models. Existing studies have explored various diversity-aware data selection methods to construct high-quality datasets and enhance model performance. However, the fundamental problem of precisely defining and measuring data diversity remains underexplored, limiting clear guidance for data engineering. To address this, we systematically analyze 11 existing diversity measurement methods by evaluating their correlation with model performance through extensive fine-tuning experiments. Our results indicate that a reliable diversity measure should properly account for both inter-sample differences and the information distribution in the sample space. Building on this, we propose NovelSum, a new diversity metric based on sample-level "novelty." Experiments on both simulated and real-world data show that NovelSum accurately captures diversity variations and achieves a 0.97 correlation with instruction-tuned model performance, highlighting its value in guiding data engineering practices. With NovelSum as an optimization objective, we further develop a greedy, diversity-oriented data selection strategy that outperforms existing approaches, validating both the effectiveness and practical significance of our metric.
Deep Generative Models Unveil Patterns in Medical Images Through Vision-Language Conditioning
Oct 17, 2024



Abstract:Deep generative models have significantly advanced medical imaging analysis by enhancing dataset size and quality. Beyond mere data augmentation, our research in this paper highlights an additional, significant capacity of deep generative models: their ability to reveal and demonstrate patterns in medical images. We employ a generative structure with hybrid conditions, combining clinical data and segmentation masks to guide the image synthesis process. Furthermore, we innovatively transformed the tabular clinical data into textual descriptions. This approach simplifies the handling of missing values and also enables us to leverage large pre-trained vision-language models that investigate the relations between independent clinical entries and comprehend general terms, such as gender and smoking status. Our approach differs from and presents a more challenging task than traditional medical report-guided synthesis due to the less visual correlation of our clinical information with the images. To overcome this, we introduce a text-visual embedding mechanism that strengthens the conditions, ensuring the network effectively utilizes the provided information. Our pipeline is generalizable to both GAN-based and diffusion models. Experiments on chest CT, particularly focusing on the smoking status, demonstrated a consistent intensity shift in the lungs which is in agreement with clinical observations, indicating the effectiveness of our method in capturing and visualizing the impact of specific attributes on medical image patterns. Our methods offer a new avenue for the early detection and precise visualization of complex clinical conditions with deep generative models. All codes are https://github.com/junzhin/DGM-VLC.
MONICA: Benchmarking on Long-tailed Medical Image Classification
Oct 02, 2024



Abstract:Long-tailed learning is considered to be an extremely challenging problem in data imbalance learning. It aims to train well-generalized models from a large number of images that follow a long-tailed class distribution. In the medical field, many diagnostic imaging exams such as dermoscopy and chest radiography yield a long-tailed distribution of complex clinical findings. Recently, long-tailed learning in medical image analysis has garnered significant attention. However, the field currently lacks a unified, strictly formulated, and comprehensive benchmark, which often leads to unfair comparisons and inconclusive results. To help the community improve the evaluation and advance, we build a unified, well-structured codebase called Medical OpeN-source Long-taIled ClassifiCAtion (MONICA), which implements over 30 methods developed in relevant fields and evaluated on 12 long-tailed medical datasets covering 6 medical domains. Our work provides valuable practical guidance and insights for the field, offering detailed analysis and discussion on the effectiveness of individual components within the inbuilt state-of-the-art methodologies. We hope this codebase serves as a comprehensive and reproducible benchmark, encouraging further advancements in long-tailed medical image learning. The codebase is publicly available on https://github.com/PyJulie/MONICA.
Coupling AI and Citizen Science in Creation of Enhanced Training Dataset for Medical Image Segmentation
Sep 04, 2024Abstract:Recent advancements in medical imaging and artificial intelligence (AI) have greatly enhanced diagnostic capabilities, but the development of effective deep learning (DL) models is still constrained by the lack of high-quality annotated datasets. The traditional manual annotation process by medical experts is time- and resource-intensive, limiting the scalability of these datasets. In this work, we introduce a robust and versatile framework that combines AI and crowdsourcing to improve both the quality and quantity of medical image datasets across different modalities. Our approach utilises a user-friendly online platform that enables a diverse group of crowd annotators to label medical images efficiently. By integrating the MedSAM segmentation AI with this platform, we accelerate the annotation process while maintaining expert-level quality through an algorithm that merges crowd-labelled images. Additionally, we employ pix2pixGAN, a generative AI model, to expand the training dataset with synthetic images that capture realistic morphological features. These methods are combined into a cohesive framework designed to produce an enhanced dataset, which can serve as a universal pre-processing pipeline to boost the training of any medical deep learning segmentation model. Our results demonstrate that this framework significantly improves model performance, especially when training data is limited.
Beyond the Hype: A dispassionate look at vision-language models in medical scenario
Aug 16, 2024Abstract:Recent advancements in Large Vision-Language Models (LVLMs) have demonstrated remarkable capabilities across diverse tasks, garnering significant attention in AI communities. However, their performance and reliability in specialized domains such as medicine remain insufficiently assessed. In particular, most assessments over-concentrate in evaluating VLMs based on simple Visual Question Answering (VQA) on multi-modality data, while ignoring the in-depth characteristic of LVLMs. In this study, we introduce RadVUQA, a novel Radiological Visual Understanding and Question Answering benchmark, to comprehensively evaluate existing LVLMs. RadVUQA mainly validates LVLMs across five dimensions: 1) Anatomical understanding, assessing the models' ability to visually identify biological structures; 2) Multimodal comprehension, which involves the capability of interpreting linguistic and visual instructions to produce desired outcomes; 3) Quantitative and spatial reasoning, evaluating the models' spatial awareness and proficiency in combining quantitative analysis with visual and linguistic information; 4) Physiological knowledge, measuring the models' capability to comprehend functions and mechanisms of organs and systems; and 5) Robustness, which assesses the models' capabilities against unharmonised and synthetic data. The results indicate that both generalized LVLMs and medical-specific LVLMs have critical deficiencies with weak multimodal comprehension and quantitative reasoning capabilities. Our findings reveal the large gap between existing LVLMs and clinicians, highlighting the urgent need for more robust and intelligent LVLMs. The code and dataset will be available after the acceptance of this paper.
Probing Perfection: The Relentless Art of Meddling for Pulmonary Airway Segmentation from HRCT via a Human-AI Collaboration Based Active Learning Method
Jul 03, 2024Abstract:In pulmonary tracheal segmentation, the scarcity of annotated data is a prevalent issue in medical segmentation. Additionally, Deep Learning (DL) methods face challenges: the opacity of 'black box' models and the need for performance enhancement. Our Human-Computer Interaction (HCI) based models (RS_UNet, LC_UNet, UUNet, and WD_UNet) address these challenges by combining diverse query strategies with various DL models. We train four HCI models and repeat these steps: (1) Query Strategy: The HCI models select samples that provide the most additional representative information when labeled in each iteration and identify unlabeled samples with the greatest predictive disparity using Wasserstein Distance, Least Confidence, Entropy Sampling, and Random Sampling. (2) Central line correction: Selected samples are used for expert correction of system-generated tracheal central lines in each training round. (3) Update training dataset: Experts update the training dataset after each DL model's training epoch, enhancing the trustworthiness and performance of the models. (4) Model training: The HCI model is trained using the updated dataset and an enhanced UNet version. Experimental results confirm the effectiveness of these HCI-based approaches, showing that WD-UNet, LC-UNet, UUNet, and RS-UNet achieve comparable or superior performance to state-of-the-art DL models. Notably, WD-UNet achieves this with only 15%-35% of the training data, reducing physician annotation time by 65%-85%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge