Giorgos Papanastasiou
Causally Steered Diffusion for Automated Video Counterfactual Generation
Jun 17, 2025Abstract:Adapting text-to-image (T2I) latent diffusion models for video editing has shown strong visual fidelity and controllability, but challenges remain in maintaining causal relationships in video content. Edits affecting causally dependent attributes risk generating unrealistic or misleading outcomes if these relationships are ignored. In this work, we propose a causally faithful framework for counterfactual video generation, guided by a vision-language model (VLM). Our method is agnostic to the underlying video editing system and does not require access to its internal mechanisms or finetuning. Instead, we guide the generation by optimizing text prompts based on an assumed causal graph, addressing the challenge of latent space control in LDMs. We evaluate our approach using standard video quality metrics and counterfactual-specific criteria, such as causal effectiveness and minimality. Our results demonstrate that causally faithful video counterfactuals can be effectively generated within the learned distribution of LDMs through prompt-based causal steering. With its compatibility with any black-box video editing system, our method holds significant potential for generating realistic "what-if" video scenarios in diverse areas such as healthcare and digital media.
A Novel Coronary Artery Registration Method Based on Super-pixel Particle Swarm Optimization
May 30, 2025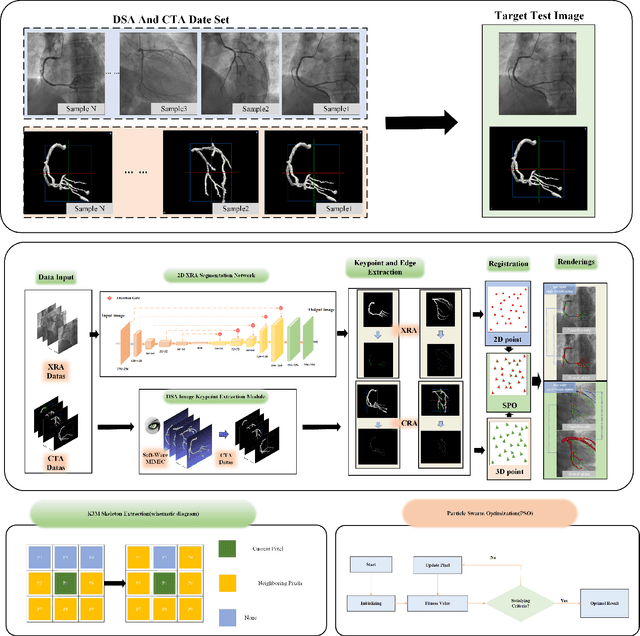
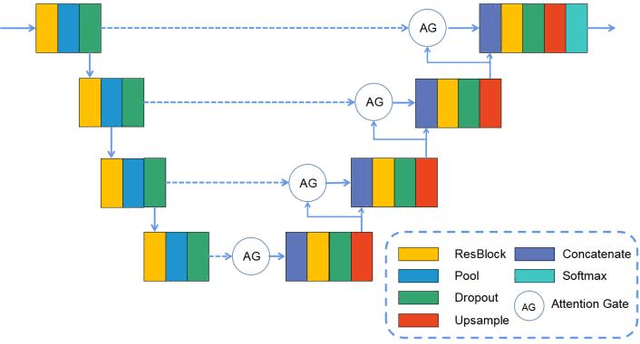
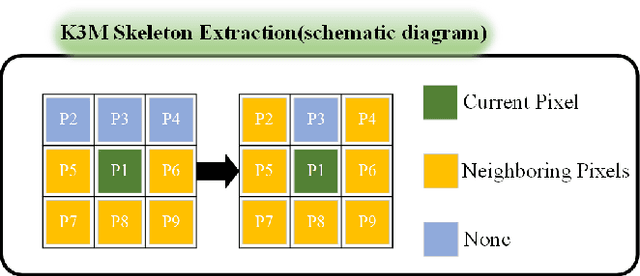
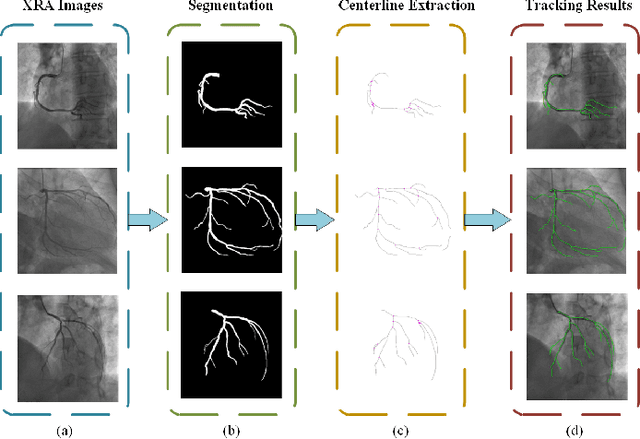
Abstract:Percutaneous Coronary Intervention (PCI) is a minimally invasive procedure that improves coronary blood flow and treats coronary artery disease. Although PCI typically requires 2D X-ray angiography (XRA) to guide catheter placement at real-time, computed tomography angiography (CTA) may substantially improve PCI by providing precise information of 3D vascular anatomy and status. To leverage real-time XRA and detailed 3D CTA anatomy for PCI, accurate multimodal image registration of XRA and CTA is required, to guide the procedure and avoid complications. This is a challenging process as it requires registration of images from different geometrical modalities (2D -> 3D and vice versa), with variations in contrast and noise levels. In this paper, we propose a novel multimodal coronary artery image registration method based on a swarm optimization algorithm, which effectively addresses challenges such as large deformations, low contrast, and noise across these imaging modalities. Our algorithm consists of two main modules: 1) preprocessing of XRA and CTA images separately, and 2) a registration module based on feature extraction using the Steger and Superpixel Particle Swarm Optimization algorithms. Our technique was evaluated on a pilot dataset of 28 pairs of XRA and CTA images from 10 patients who underwent PCI. The algorithm was compared with four state-of-the-art (SOTA) methods in terms of registration accuracy, robustness, and efficiency. Our method outperformed the selected SOTA baselines in all aspects. Experimental results demonstrate the significant effectiveness of our algorithm, surpassing the previous benchmarks and proposes a novel clinical approach that can potentially have merit for improving patient outcomes in coronary artery disease.
Reason Like a Radiologist: Chain-of-Thought and Reinforcement Learning for Verifiable Report Generation
Apr 25, 2025Abstract:Radiology report generation is critical for efficiency but current models lack the structured reasoning of experts, hindering clinical trust and explainability by failing to link visual findings to precise anatomical locations. This paper introduces BoxMed-RL, a groundbreaking unified training framework for generating spatially verifiable and explainable radiology reports. Built on a large vision-language model, BoxMed-RL revolutionizes report generation through two integrated phases: (1) In the Pretraining Phase, we refine the model via medical concept learning, using Chain-of-Thought supervision to internalize the radiologist-like workflow, followed by spatially verifiable reinforcement, which applies reinforcement learning to align medical findings with bounding boxes. (2) In the Downstream Adapter Phase, we freeze the pretrained weights and train a downstream adapter to ensure fluent and clinically credible reports. This framework precisely mimics radiologists' workflow, compelling the model to connect high-level medical concepts with definitive anatomical evidence. Extensive experiments on public datasets demonstrate that BoxMed-RL achieves an average 7% improvement in both METEOR and ROUGE-L metrics compared to state-of-the-art methods. An average 5% improvement in large language model-based metrics further underscores BoxMed-RL's robustness in generating high-quality radiology reports.
Revisiting Medical Image Retrieval via Knowledge Consolidation
Mar 12, 2025Abstract:As artificial intelligence and digital medicine increasingly permeate healthcare systems, robust governance frameworks are essential to ensure ethical, secure, and effective implementation. In this context, medical image retrieval becomes a critical component of clinical data management, playing a vital role in decision-making and safeguarding patient information. Existing methods usually learn hash functions using bottleneck features, which fail to produce representative hash codes from blended embeddings. Although contrastive hashing has shown superior performance, current approaches often treat image retrieval as a classification task, using category labels to create positive/negative pairs. Moreover, many methods fail to address the out-of-distribution (OOD) issue when models encounter external OOD queries or adversarial attacks. In this work, we propose a novel method to consolidate knowledge of hierarchical features and optimisation functions. We formulate the knowledge consolidation by introducing Depth-aware Representation Fusion (DaRF) and Structure-aware Contrastive Hashing (SCH). DaRF adaptively integrates shallow and deep representations into blended features, and SCH incorporates image fingerprints to enhance the adaptability of positive/negative pairings. These blended features further facilitate OOD detection and content-based recommendation, contributing to a secure AI-driven healthcare environment. Moreover, we present a content-guided ranking to improve the robustness and reproducibility of retrieval results. Our comprehensive assessments demonstrate that the proposed method could effectively recognise OOD samples and significantly outperform existing approaches in medical image retrieval (p<0.05). In particular, our method achieves a 5.6-38.9% improvement in mean Average Precision on the anatomical radiology dataset.
Anatomy-Guided Radiology Report Generation with Pathology-Aware Regional Prompts
Nov 16, 2024



Abstract:Radiology reporting generative AI holds significant potential to alleviate clinical workloads and streamline medical care. However, achieving high clinical accuracy is challenging, as radiological images often feature subtle lesions and intricate structures. Existing systems often fall short, largely due to their reliance on fixed size, patch-level image features and insufficient incorporation of pathological information. This can result in the neglect of such subtle patterns and inconsistent descriptions of crucial pathologies. To address these challenges, we propose an innovative approach that leverages pathology-aware regional prompts to explicitly integrate anatomical and pathological information of various scales, significantly enhancing the precision and clinical relevance of generated reports. We develop an anatomical region detector that extracts features from distinct anatomical areas, coupled with a novel multi-label lesion detector that identifies global pathologies. Our approach emulates the diagnostic process of radiologists, producing clinically accurate reports with comprehensive diagnostic capabilities. Experimental results show that our model outperforms previous state-of-the-art methods on most natural language generation and clinical efficacy metrics, with formal expert evaluations affirming its potential to enhance radiology practice.
SeLoRA: Self-Expanding Low-Rank Adaptation of Latent Diffusion Model for Medical Image Synthesis
Aug 13, 2024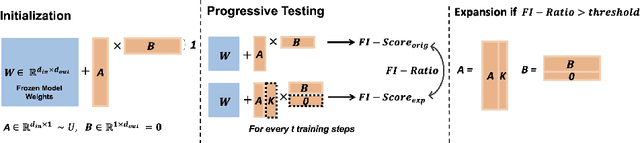
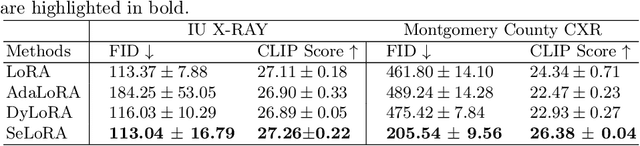
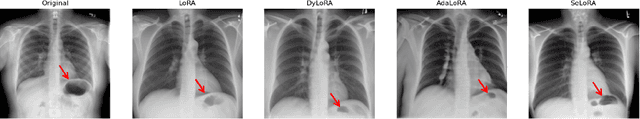

Abstract:The persistent challenge of medical image synthesis posed by the scarcity of annotated data and the need to synthesize `missing modalities' for multi-modal analysis, underscored the imperative development of effective synthesis methods. Recently, the combination of Low-Rank Adaptation (LoRA) with latent diffusion models (LDMs) has emerged as a viable approach for efficiently adapting pre-trained large language models, in the medical field. However, the direct application of LoRA assumes uniform ranking across all linear layers, overlooking the significance of different weight matrices, and leading to sub-optimal outcomes. Prior works on LoRA prioritize the reduction of trainable parameters, and there exists an opportunity to further tailor this adaptation process to the intricate demands of medical image synthesis. In response, we present SeLoRA, a Self-Expanding Low-Rank Adaptation Module, that dynamically expands its ranking across layers during training, strategically placing additional ranks on crucial layers, to allow the model to elevate synthesis quality where it matters most. The proposed method not only enables LDMs to fine-tune on medical data efficiently but also empowers the model to achieve improved image quality with minimal ranking. The code of our SeLoRA method is publicly available on https://anonymous.4open.science/r/SeLoRA-980D .
Advancing oncology with federated learning: transcending boundaries in breast, lung, and prostate cancer. A systematic review
Aug 08, 2024Abstract:Federated Learning (FL) has emerged as a promising solution to address the limitations of centralised machine learning (ML) in oncology, particularly in overcoming privacy concerns and harnessing the power of diverse, multi-center data. This systematic review synthesises current knowledge on the state-of-the-art FL in oncology, focusing on breast, lung, and prostate cancer. Distinct from previous surveys, our comprehensive review critically evaluates the real-world implementation and impact of FL on cancer care, demonstrating its effectiveness in enhancing ML generalisability, performance and data privacy in clinical settings and data. We evaluated state-of-the-art advances in FL, demonstrating its growing adoption amid tightening data privacy regulations. FL outperformed centralised ML in 15 out of the 25 studies reviewed, spanning diverse ML models and clinical applications, and facilitating integration of multi-modal information for precision medicine. Despite the current challenges identified in reproducibility, standardisation and methodology across studies, the demonstrable benefits of FL in harnessing real-world data and addressing clinical needs highlight its significant potential for advancing cancer research. We propose that future research should focus on addressing these limitations and investigating further advanced FL methods, to fully harness data diversity and realise the transformative power of cutting-edge FL in cancer care.
Artificial Immunofluorescence in a Flash: Rapid Synthetic Imaging from Brightfield Through Residual Diffusion
Jul 25, 2024



Abstract:Immunofluorescent (IF) imaging is crucial for visualizing biomarker expressions, cell morphology and assessing the effects of drug treatments on sub-cellular components. IF imaging needs extra staining process and often requiring cell fixation, therefore it may also introduce artefects and alter endogenouous cell morphology. Some IF stains are expensive or not readily available hence hindering experiments. Recent diffusion models, which synthesise high-fidelity IF images from easy-to-acquire brightfield (BF) images, offer a promising solution but are hindered by training instability and slow inference times due to the noise diffusion process. This paper presents a novel method for the conditional synthesis of IF images directly from BF images along with cell segmentation masks. Our approach employs a Residual Diffusion process that enhances stability and significantly reduces inference time. We performed a critical evaluation against other image-to-image synthesis models, including UNets, GANs, and advanced diffusion models. Our model demonstrates significant improvements in image quality (p<0.05 in MSE, PSNR, and SSIM), inference speed (26 times faster than competing diffusion models), and accurate segmentation results for both nuclei and cell bodies (0.77 and 0.63 mean IOU for nuclei and cell true positives, respectively). This paper is a substantial advancement in the field, providing robust and efficient tools for cell image analysis.
Benchmarking Counterfactual Image Generation
Mar 29, 2024



Abstract:Counterfactual image generation is pivotal for understanding the causal relations of variables, with applications in interpretability and generation of unbiased synthetic data. However, evaluating image generation is a long-standing challenge in itself. The need to evaluate counterfactual generation compounds on this challenge, precisely because counterfactuals, by definition, are hypothetical scenarios without observable ground truths. In this paper, we present a novel comprehensive framework aimed at benchmarking counterfactual image generation methods. We incorporate metrics that focus on evaluating diverse aspects of counterfactuals, such as composition, effectiveness, minimality of interventions, and image realism. We assess the performance of three distinct conditional image generation model types, based on the Structural Causal Model paradigm. Our work is accompanied by a user-friendly Python package which allows to further evaluate and benchmark existing and future counterfactual image generation methods. Our framework is extendable to additional SCM and other causal methods, generative models, and datasets.
From explainable to interpretable deep learning for natural language processing in healthcare: how far from reality?
Mar 18, 2024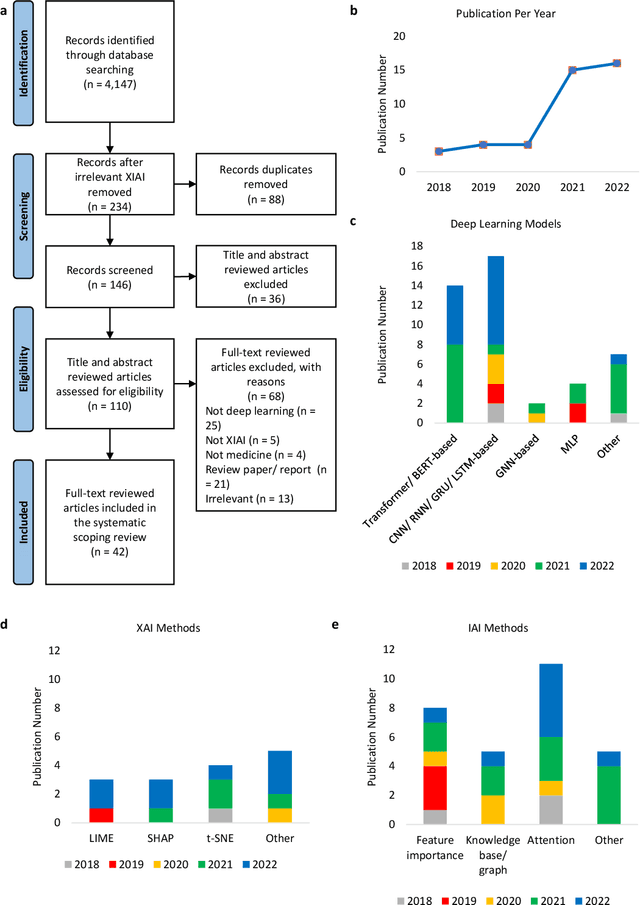
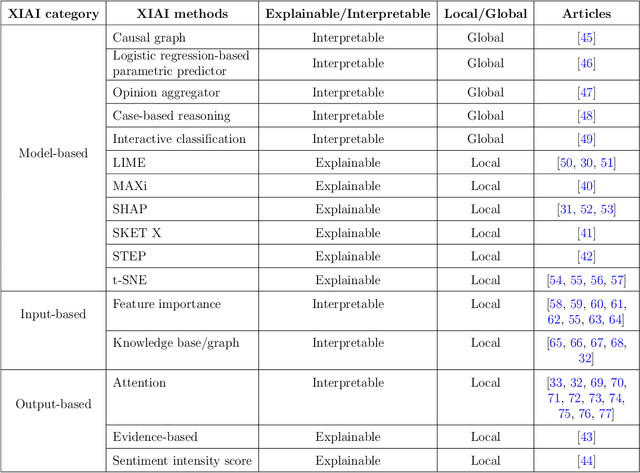
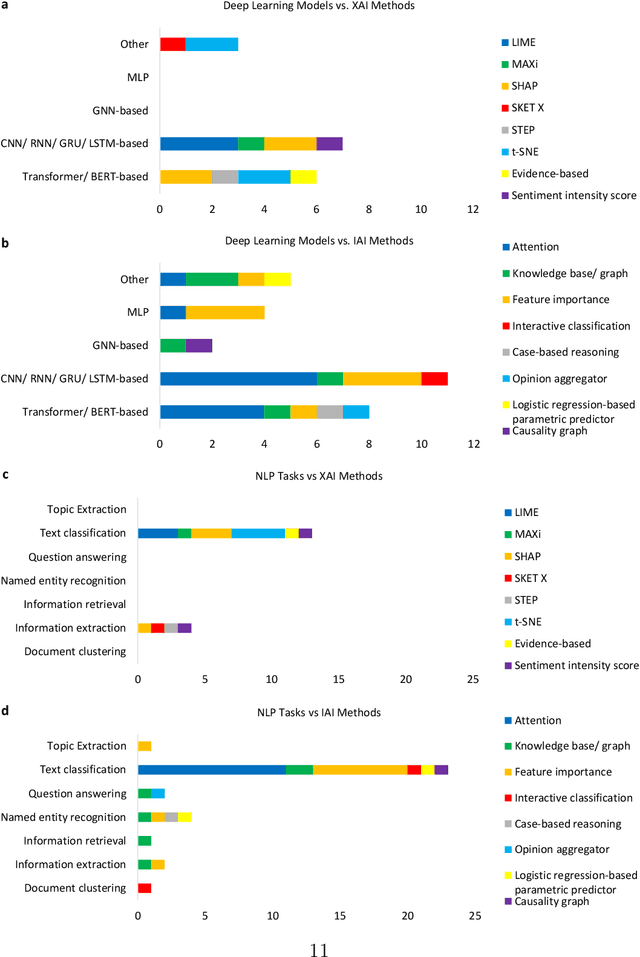
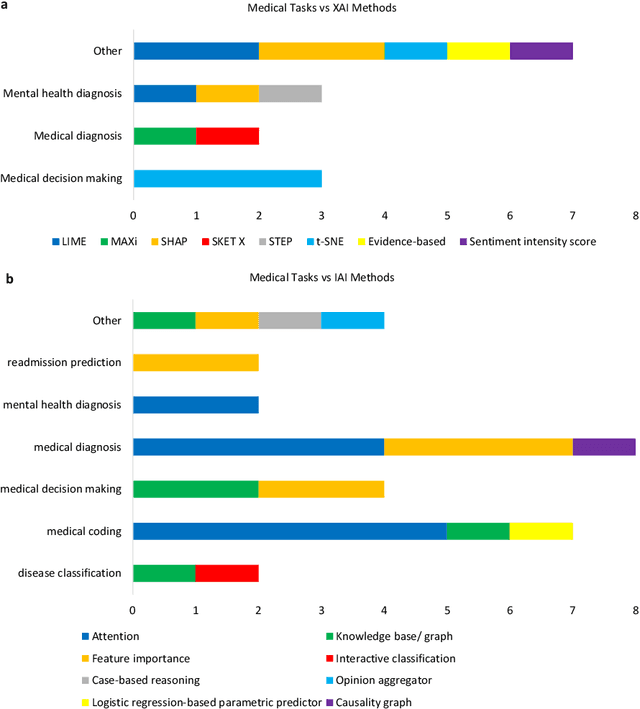
Abstract:Deep learning (DL) has substantially enhanced healthcare research by addressing various natural language processing (NLP) tasks. Yet, the increasing complexity of DL-based NLP methods necessitates transparent model interpretability, or at least explainability, for reliable decision-making. This work presents a thorough scoping review on explainable and interpretable DL in healthcare NLP. The term "XIAI" (eXplainable and Interpretable Artificial Intelligence) was introduced to distinguish XAI from IAI. Methods were further categorized based on their functionality (model-, input-, output-based) and scope (local, global). Our analysis shows that attention mechanisms were the most dominant emerging IAI. Moreover, IAI is increasingly used against XAI. The major challenges identified are that most XIAI do not explore "global" modeling processes, the lack of best practices, and the unmet need for systematic evaluation and benchmarks. Important opportunities were raised such as using "attention" to enhance multi-modal XIAI for personalized medicine and combine DL with causal reasoning. Our discussion encourages the integration of XIAI in LLMs and domain-specific smaller models. Our review can stimulate further research and benchmarks toward improving inherent IAI and engaging complex NLP in healthcare.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge