Matthieu Komorowski
Faculty of Medicine, Department of Surgery and Cancer, Imperial College London, London, United Kingdom
Unpaired Translation of Chest X-ray Images for Lung Opacity Diagnosis via Adaptive Activation Masks and Cross-Domain Alignment
Mar 25, 2025Abstract:Chest X-ray radiographs (CXRs) play a pivotal role in diagnosing and monitoring cardiopulmonary diseases. However, lung opac- ities in CXRs frequently obscure anatomical structures, impeding clear identification of lung borders and complicating the localization of pathology. This challenge significantly hampers segmentation accuracy and precise lesion identification, which are crucial for diagnosis. To tackle these issues, our study proposes an unpaired CXR translation framework that converts CXRs with lung opacities into counterparts without lung opacities while preserving semantic features. Central to our approach is the use of adaptive activation masks to selectively modify opacity regions in lung CXRs. Cross-domain alignment ensures translated CXRs without opacity issues align with feature maps and prediction labels from a pre-trained CXR lesion classifier, facilitating the interpretability of the translation process. We validate our method using RSNA, MIMIC-CXR-JPG and JSRT datasets, demonstrating superior translation quality through lower Frechet Inception Distance (FID) and Kernel Inception Distance (KID) scores compared to existing meth- ods (FID: 67.18 vs. 210.4, KID: 0.01604 vs. 0.225). Evaluation on RSNA opacity, MIMIC acute respiratory distress syndrome (ARDS) patient CXRs and JSRT CXRs show our method enhances segmentation accuracy of lung borders and improves lesion classification, further underscoring its potential in clinical settings (RSNA: mIoU: 76.58% vs. 62.58%, Sensitivity: 85.58% vs. 77.03%; MIMIC ARDS: mIoU: 86.20% vs. 72.07%, Sensitivity: 92.68% vs. 86.85%; JSRT: mIoU: 91.08% vs. 85.6%, Sensitivity: 97.62% vs. 95.04%). Our approach advances CXR imaging analysis, especially in investigating segmentation impacts through image translation techniques.
Anatomy-Guided Radiology Report Generation with Pathology-Aware Regional Prompts
Nov 16, 2024



Abstract:Radiology reporting generative AI holds significant potential to alleviate clinical workloads and streamline medical care. However, achieving high clinical accuracy is challenging, as radiological images often feature subtle lesions and intricate structures. Existing systems often fall short, largely due to their reliance on fixed size, patch-level image features and insufficient incorporation of pathological information. This can result in the neglect of such subtle patterns and inconsistent descriptions of crucial pathologies. To address these challenges, we propose an innovative approach that leverages pathology-aware regional prompts to explicitly integrate anatomical and pathological information of various scales, significantly enhancing the precision and clinical relevance of generated reports. We develop an anatomical region detector that extracts features from distinct anatomical areas, coupled with a novel multi-label lesion detector that identifies global pathologies. Our approach emulates the diagnostic process of radiologists, producing clinically accurate reports with comprehensive diagnostic capabilities. Experimental results show that our model outperforms previous state-of-the-art methods on most natural language generation and clinical efficacy metrics, with formal expert evaluations affirming its potential to enhance radiology practice.
Multi-site, Multi-domain Airway Tree Modeling : A Public Benchmark for Pulmonary Airway Segmentation
Mar 10, 2023



Abstract:Open international challenges are becoming the de facto standard for assessing computer vision and image analysis algorithms. In recent years, new methods have extended the reach of pulmonary airway segmentation that is closer to the limit of image resolution. Since EXACT'09 pulmonary airway segmentation, limited effort has been directed to quantitative comparison of newly emerged algorithms driven by the maturity of deep learning based approaches and clinical drive for resolving finer details of distal airways for early intervention of pulmonary diseases. Thus far, public annotated datasets are extremely limited, hindering the development of data-driven methods and detailed performance evaluation of new algorithms. To provide a benchmark for the medical imaging community, we organized the Multi-site, Multi-domain Airway Tree Modeling (ATM'22), which was held as an official challenge event during the MICCAI 2022 conference. ATM'22 provides large-scale CT scans with detailed pulmonary airway annotation, including 500 CT scans (300 for training, 50 for validation, and 150 for testing). The dataset was collected from different sites and it further included a portion of noisy COVID-19 CTs with ground-glass opacity and consolidation. Twenty-three teams participated in the entire phase of the challenge and the algorithms for the top ten teams are reviewed in this paper. Quantitative and qualitative results revealed that deep learning models embedded with the topological continuity enhancement achieved superior performance in general. ATM'22 challenge holds as an open-call design, the training data and the gold standard evaluation are available upon successful registration via its homepage.
Beyond Low Earth Orbit: Biomonitoring, Artificial Intelligence, and Precision Space Health
Dec 22, 2021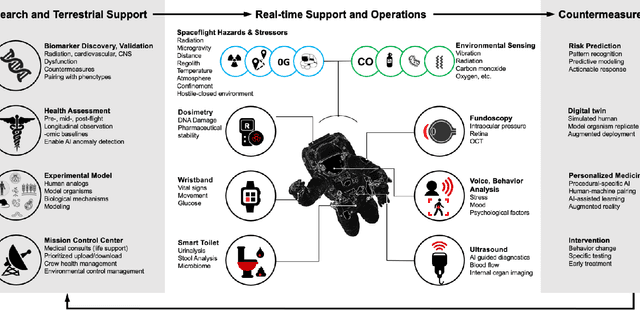
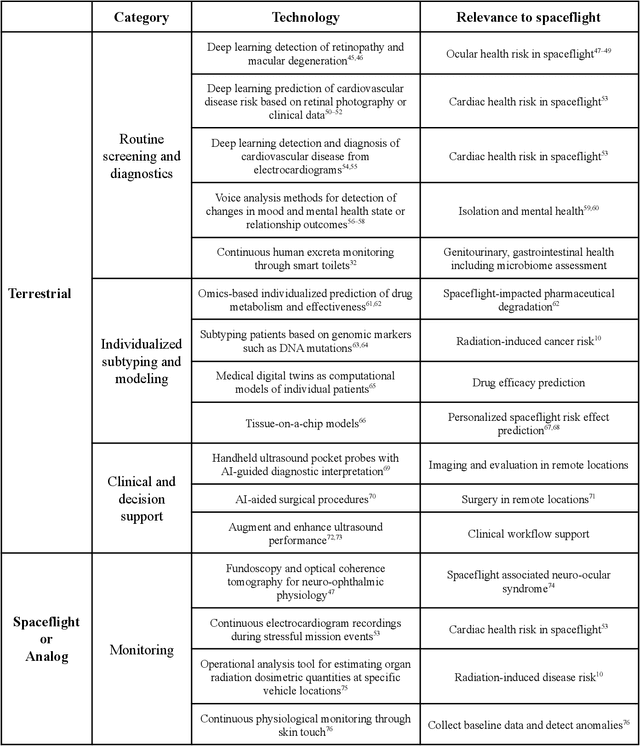
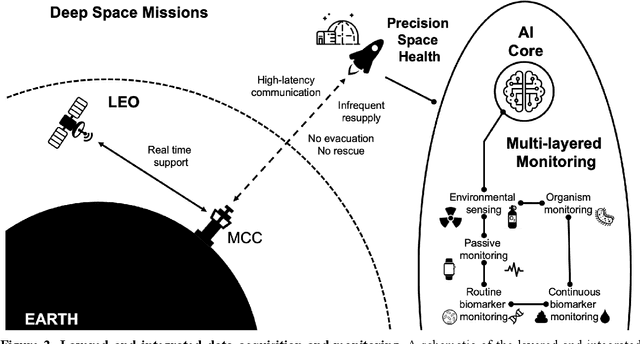
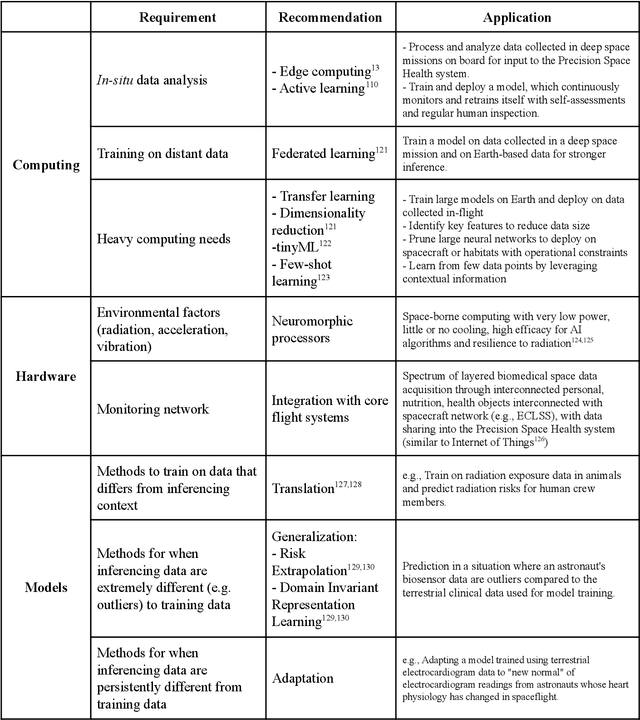
Abstract:Human space exploration beyond low Earth orbit will involve missions of significant distance and duration. To effectively mitigate myriad space health hazards, paradigm shifts in data and space health systems are necessary to enable Earth-independence, rather than Earth-reliance. Promising developments in the fields of artificial intelligence and machine learning for biology and health can address these needs. We propose an appropriately autonomous and intelligent Precision Space Health system that will monitor, aggregate, and assess biomedical statuses; analyze and predict personalized adverse health outcomes; adapt and respond to newly accumulated data; and provide preventive, actionable, and timely insights to individual deep space crew members and iterative decision support to their crew medical officer. Here we present a summary of recommendations from a workshop organized by the National Aeronautics and Space Administration, on future applications of artificial intelligence in space biology and health. In the next decade, biomonitoring technology, biomarker science, spacecraft hardware, intelligent software, and streamlined data management must mature and be woven together into a Precision Space Health system to enable humanity to thrive in deep space.
Beyond Low Earth Orbit: Biological Research, Artificial Intelligence, and Self-Driving Labs
Dec 22, 2021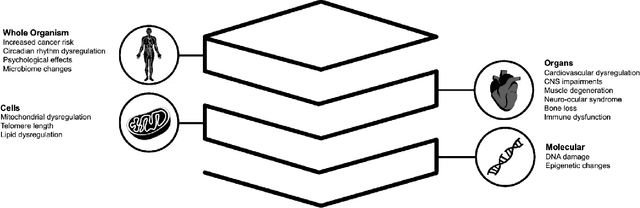
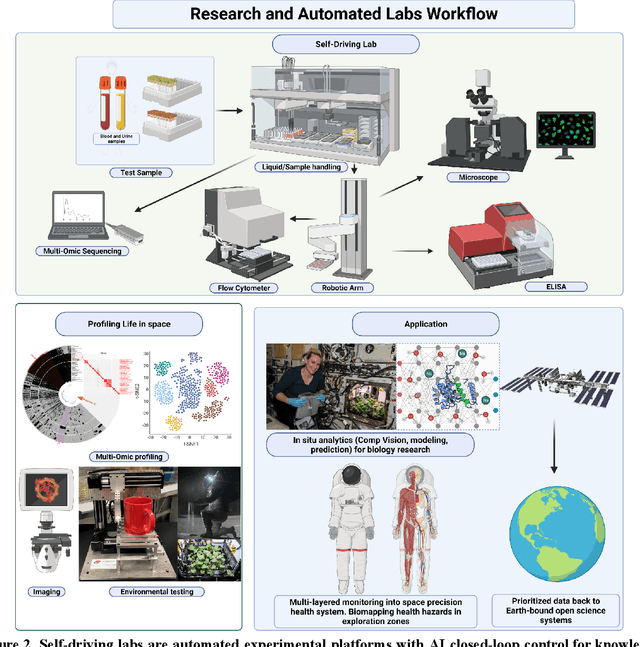
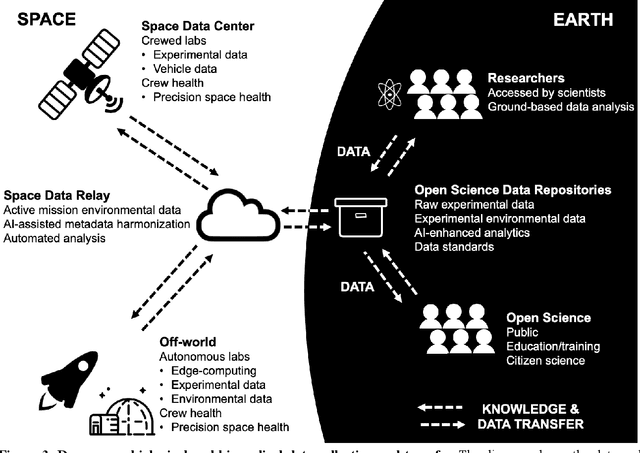
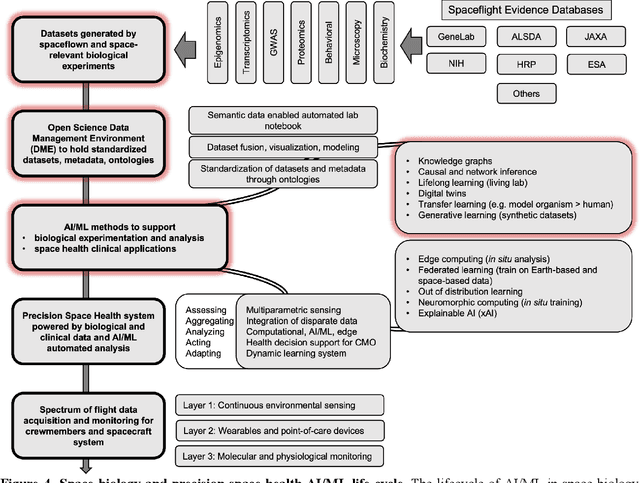
Abstract:Space biology research aims to understand fundamental effects of spaceflight on organisms, develop foundational knowledge to support deep space exploration, and ultimately bioengineer spacecraft and habitats to stabilize the ecosystem of plants, crops, microbes, animals, and humans for sustained multi-planetary life. To advance these aims, the field leverages experiments, platforms, data, and model organisms from both spaceborne and ground-analog studies. As research is extended beyond low Earth orbit, experiments and platforms must be maximally autonomous, light, agile, and intelligent to expedite knowledge discovery. Here we present a summary of recommendations from a workshop organized by the National Aeronautics and Space Administration on artificial intelligence, machine learning, and modeling applications which offer key solutions toward these space biology challenges. In the next decade, the synthesis of artificial intelligence into the field of space biology will deepen the biological understanding of spaceflight effects, facilitate predictive modeling and analytics, support maximally autonomous and reproducible experiments, and efficiently manage spaceborne data and metadata, all with the goal to enable life to thrive in deep space.
Enabling risk-aware Reinforcement Learning for medical interventions through uncertainty decomposition
Sep 16, 2021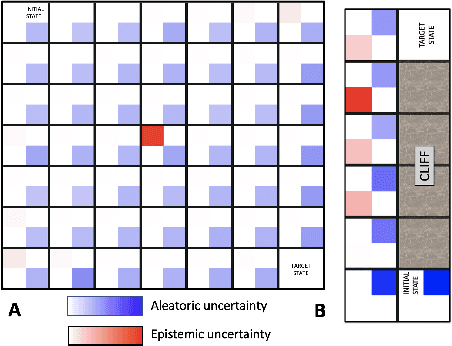

Abstract:Reinforcement Learning (RL) is emerging as tool for tackling complex control and decision-making problems. However, in high-risk environments such as healthcare, manufacturing, automotive or aerospace, it is often challenging to bridge the gap between an apparently optimal policy learnt by an agent and its real-world deployment, due to the uncertainties and risk associated with it. Broadly speaking RL agents face two kinds of uncertainty, 1. aleatoric uncertainty, which reflects randomness or noise in the dynamics of the world, and 2. epistemic uncertainty, which reflects the bounded knowledge of the agent due to model limitations and finite amount of information/data the agent has acquired about the world. These two types of uncertainty carry fundamentally different implications for the evaluation of performance and the level of risk or trust. Yet these aleatoric and epistemic uncertainties are generally confounded as standard and even distributional RL is agnostic to this difference. Here we propose how a distributional approach (UA-DQN) can be recast to render uncertainties by decomposing the net effects of each uncertainty. We demonstrate the operation of this method in grid world examples to build intuition and then show a proof of concept application for an RL agent operating as a clinical decision support system in critical care
Optimizing Sequential Medical Treatments with Auto-Encoding Heuristic Search in POMDPs
May 17, 2019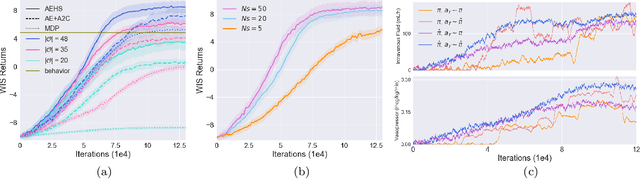
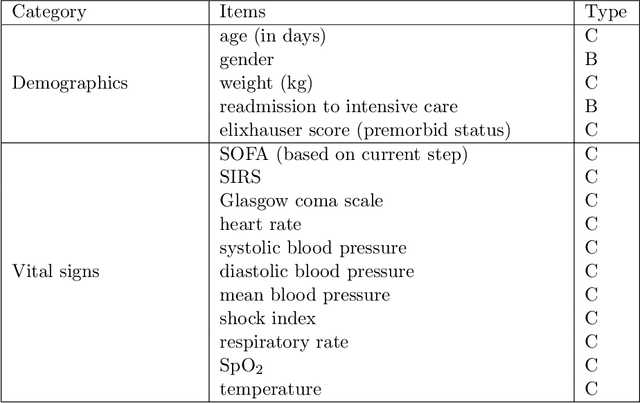
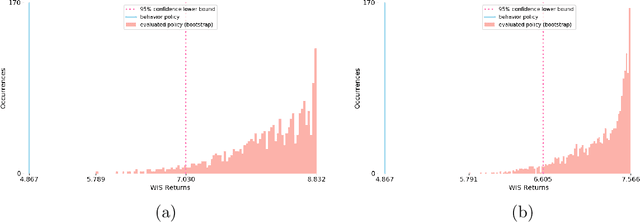
Abstract:Health-related data is noisy and stochastic in implying the true physiological states of patients, limiting information contained in single-moment observations for sequential clinical decision making. We model patient-clinician interactions as partially observable Markov decision processes (POMDPs) and optimize sequential treatment based on belief states inferred from history sequence. To facilitate inference, we build a variational generative model and boost state representation with a recurrent neural network (RNN), incorporating an auxiliary loss from sequence auto-encoding. Meanwhile, we optimize a continuous policy of drug levels with an actor-critic method where policy gradients are obtained from a stablized off-policy estimate of advantage function, with the value of belief state backed up by parallel best-first suffix trees. We exploit our methodology in optimizing dosages of vasopressor and intravenous fluid for sepsis patients using a retrospective intensive care dataset and evaluate the learned policy with off-policy policy evaluation (OPPE). The results demonstrate that modelling as POMDPs yields better performance than MDPs, and that incorporating heuristic search improves sample efficiency.
Understanding the Artificial Intelligence Clinician and optimal treatment strategies for sepsis in intensive care
Mar 06, 2019Abstract:In this document, we explore in more detail our published work (Komorowski, Celi, Badawi, Gordon, & Faisal, 2018) for the benefit of the AI in Healthcare research community. In the above paper, we developed the AI Clinician system, which demonstrated how reinforcement learning could be used to make useful recommendations towards optimal treatment decisions from intensive care data. Since publication a number of authors have reviewed our work (e.g. Abbasi, 2018; Bos, Azoulay, & Martin-Loeches, 2019; Saria, 2018). Given the difference of our framework to previous work, the fact that we are bridging two very different academic communities (intensive care and machine learning) and that our work has impact on a number of other areas with more traditional computer-based approaches (biosignal processing and control, biomedical engineering), we are providing here additional details on our recent publication.
Improving Sepsis Treatment Strategies by Combining Deep and Kernel-Based Reinforcement Learning
Jan 15, 2019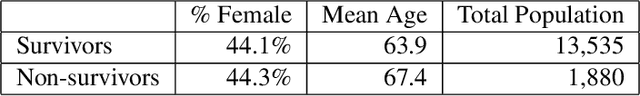
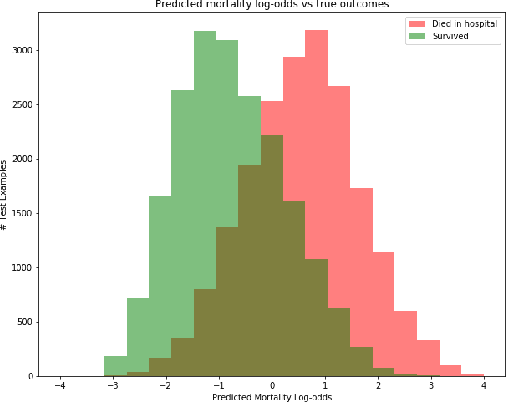

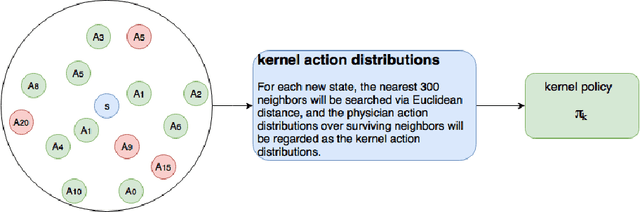
Abstract:Sepsis is the leading cause of mortality in the ICU. It is challenging to manage because individual patients respond differently to treatment. Thus, tailoring treatment to the individual patient is essential for the best outcomes. In this paper, we take steps toward this goal by applying a mixture-of-experts framework to personalize sepsis treatment. The mixture model selectively alternates between neighbor-based (kernel) and deep reinforcement learning (DRL) experts depending on patient's current history. On a large retrospective cohort, this mixture-based approach outperforms physician, kernel only, and DRL-only experts.
Model-Based Reinforcement Learning for Sepsis Treatment
Nov 23, 2018
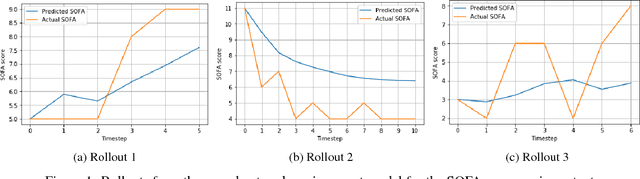

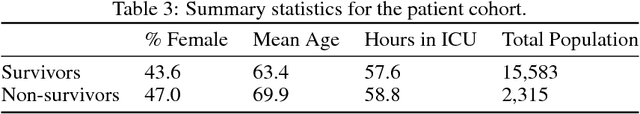
Abstract:Sepsis is a dangerous condition that is a leading cause of patient mortality. Treating sepsis is highly challenging, because individual patients respond very differently to medical interventions and there is no universally agreed-upon treatment for sepsis. In this work, we explore the use of continuous state-space model-based reinforcement learning (RL) to discover high-quality treatment policies for sepsis patients. Our quantitative evaluation reveals that by blending the treatment strategy discovered with RL with what clinicians follow, we can obtain improved policies, potentially allowing for better medical treatment for sepsis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge