Yingying Fang
Reason Like a Radiologist: Chain-of-Thought and Reinforcement Learning for Verifiable Report Generation
Apr 25, 2025Abstract:Radiology report generation is critical for efficiency but current models lack the structured reasoning of experts, hindering clinical trust and explainability by failing to link visual findings to precise anatomical locations. This paper introduces BoxMed-RL, a groundbreaking unified training framework for generating spatially verifiable and explainable radiology reports. Built on a large vision-language model, BoxMed-RL revolutionizes report generation through two integrated phases: (1) In the Pretraining Phase, we refine the model via medical concept learning, using Chain-of-Thought supervision to internalize the radiologist-like workflow, followed by spatially verifiable reinforcement, which applies reinforcement learning to align medical findings with bounding boxes. (2) In the Downstream Adapter Phase, we freeze the pretrained weights and train a downstream adapter to ensure fluent and clinically credible reports. This framework precisely mimics radiologists' workflow, compelling the model to connect high-level medical concepts with definitive anatomical evidence. Extensive experiments on public datasets demonstrate that BoxMed-RL achieves an average 7% improvement in both METEOR and ROUGE-L metrics compared to state-of-the-art methods. An average 5% improvement in large language model-based metrics further underscores BoxMed-RL's robustness in generating high-quality radiology reports.
Unpaired Translation of Chest X-ray Images for Lung Opacity Diagnosis via Adaptive Activation Masks and Cross-Domain Alignment
Mar 25, 2025Abstract:Chest X-ray radiographs (CXRs) play a pivotal role in diagnosing and monitoring cardiopulmonary diseases. However, lung opac- ities in CXRs frequently obscure anatomical structures, impeding clear identification of lung borders and complicating the localization of pathology. This challenge significantly hampers segmentation accuracy and precise lesion identification, which are crucial for diagnosis. To tackle these issues, our study proposes an unpaired CXR translation framework that converts CXRs with lung opacities into counterparts without lung opacities while preserving semantic features. Central to our approach is the use of adaptive activation masks to selectively modify opacity regions in lung CXRs. Cross-domain alignment ensures translated CXRs without opacity issues align with feature maps and prediction labels from a pre-trained CXR lesion classifier, facilitating the interpretability of the translation process. We validate our method using RSNA, MIMIC-CXR-JPG and JSRT datasets, demonstrating superior translation quality through lower Frechet Inception Distance (FID) and Kernel Inception Distance (KID) scores compared to existing meth- ods (FID: 67.18 vs. 210.4, KID: 0.01604 vs. 0.225). Evaluation on RSNA opacity, MIMIC acute respiratory distress syndrome (ARDS) patient CXRs and JSRT CXRs show our method enhances segmentation accuracy of lung borders and improves lesion classification, further underscoring its potential in clinical settings (RSNA: mIoU: 76.58% vs. 62.58%, Sensitivity: 85.58% vs. 77.03%; MIMIC ARDS: mIoU: 86.20% vs. 72.07%, Sensitivity: 92.68% vs. 86.85%; JSRT: mIoU: 91.08% vs. 85.6%, Sensitivity: 97.62% vs. 95.04%). Our approach advances CXR imaging analysis, especially in investigating segmentation impacts through image translation techniques.
GEMA-Score: Granular Explainable Multi-Agent Score for Radiology Report Evaluation
Mar 07, 2025



Abstract:Automatic medical report generation supports clinical diagnosis, reduces the workload of radiologists, and holds the promise of improving diagnosis consistency. However, existing evaluation metrics primarily assess the accuracy of key medical information coverage in generated reports compared to human-written reports, while overlooking crucial details such as the location and certainty of reported abnormalities. These limitations hinder the comprehensive assessment of the reliability of generated reports and pose risks in their selection for clinical use. Therefore, we propose a Granular Explainable Multi-Agent Score (GEMA-Score) in this paper, which conducts both objective quantification and subjective evaluation through a large language model-based multi-agent workflow. Our GEMA-Score parses structured reports and employs NER-F1 calculations through interactive exchanges of information among agents to assess disease diagnosis, location, severity, and uncertainty. Additionally, an LLM-based scoring agent evaluates completeness, readability, and clinical terminology while providing explanatory feedback. Extensive experiments validate that GEMA-Score achieves the highest correlation with human expert evaluations on a public dataset, demonstrating its effectiveness in clinical scoring (Kendall coefficient = 0.70 for Rexval dataset and Kendall coefficient = 0.54 for RadEvalX dataset). The anonymous project demo is available at: https://github.com/Zhenxuan-Zhang/GEMA_score.
Decoding Report Generators: A Cyclic Vision-Language Adapter for Counterfactual Explanations
Nov 08, 2024
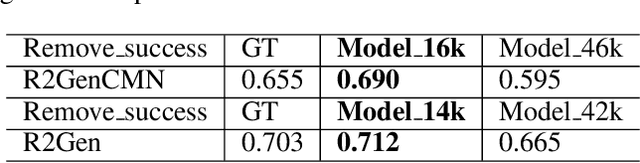
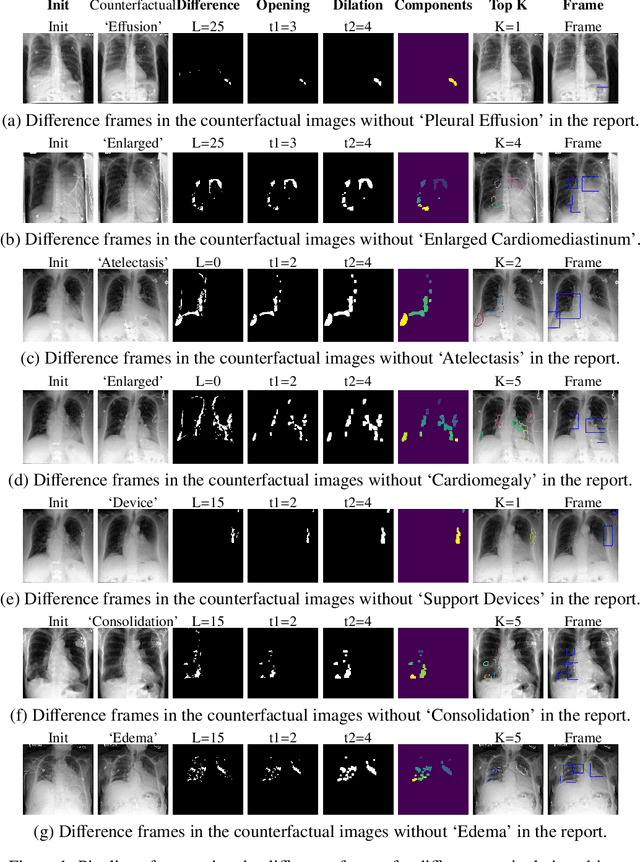
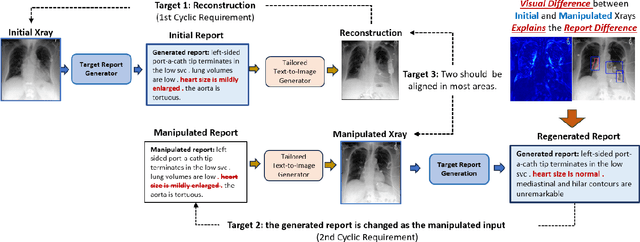
Abstract:Despite significant advancements in report generation methods, a critical limitation remains: the lack of interpretability in the generated text. This paper introduces an innovative approach to enhance the explainability of text generated by report generation models. Our method employs cyclic text manipulation and visual comparison to identify and elucidate the features in the original content that influence the generated text. By manipulating the generated reports and producing corresponding images, we create a comparative framework that highlights key attributes and their impact on the text generation process. This approach not only identifies the image features aligned to the generated text but also improves transparency but also provides deeper insights into the decision-making mechanisms of the report generation models. Our findings demonstrate the potential of this method to significantly enhance the interpretability and transparency of AI-generated reports.
Learning Task-Specific Sampling Strategy for Sparse-View CT Reconstruction
Sep 03, 2024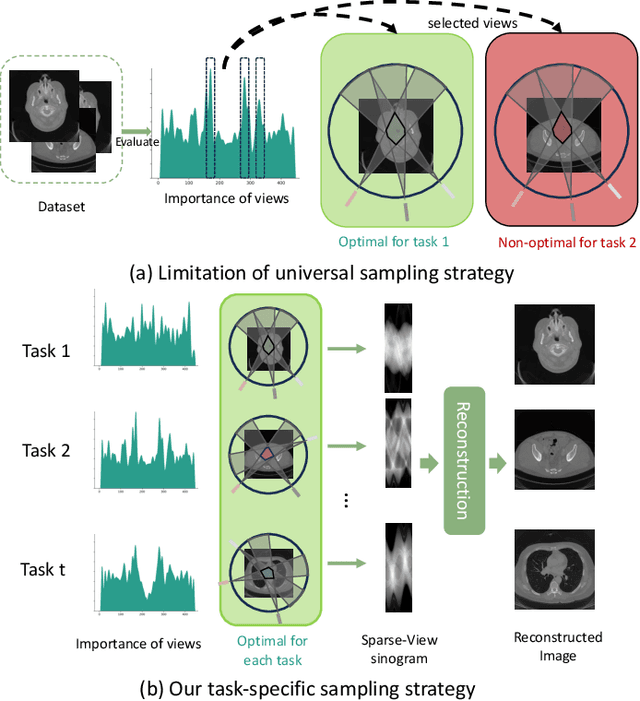
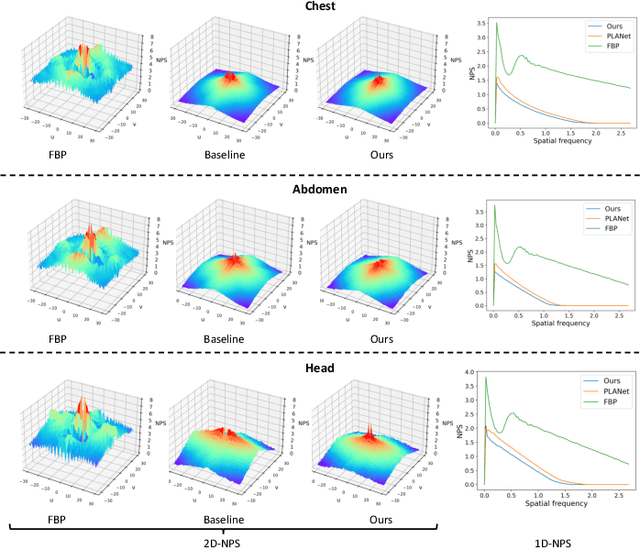

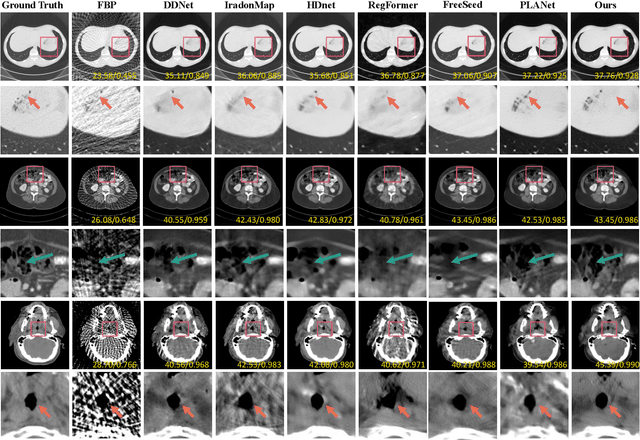
Abstract:Sparse-View Computed Tomography (SVCT) offers low-dose and fast imaging but suffers from severe artifacts. Optimizing the sampling strategy is an essential approach to improving the imaging quality of SVCT. However, current methods typically optimize a universal sampling strategy for all types of scans, overlooking the fact that the optimal strategy may vary depending on the specific scanning task, whether it involves particular body scans (e.g., chest CT scans) or downstream clinical applications (e.g., disease diagnosis). The optimal strategy for one scanning task may not perform as well when applied to other tasks. To address this problem, we propose a deep learning framework that learns task-specific sampling strategies with a multi-task approach to train a unified reconstruction network while tailoring optimal sampling strategies for each individual task. Thus, a task-specific sampling strategy can be applied for each type of scans to improve the quality of SVCT imaging and further assist in performance of downstream clinical usage. Extensive experiments across different scanning types provide validation for the effectiveness of task-specific sampling strategies in enhancing imaging quality. Experiments involving downstream tasks verify the clinical value of learned sampling strategies, as evidenced by notable improvements in downstream task performance. Furthermore, the utilization of a multi-task framework with a shared reconstruction network facilitates deployment on current imaging devices with switchable task-specific modules, and allows for easily integrate new tasks without retraining the entire model.
Probing Perfection: The Relentless Art of Meddling for Pulmonary Airway Segmentation from HRCT via a Human-AI Collaboration Based Active Learning Method
Jul 03, 2024Abstract:In pulmonary tracheal segmentation, the scarcity of annotated data is a prevalent issue in medical segmentation. Additionally, Deep Learning (DL) methods face challenges: the opacity of 'black box' models and the need for performance enhancement. Our Human-Computer Interaction (HCI) based models (RS_UNet, LC_UNet, UUNet, and WD_UNet) address these challenges by combining diverse query strategies with various DL models. We train four HCI models and repeat these steps: (1) Query Strategy: The HCI models select samples that provide the most additional representative information when labeled in each iteration and identify unlabeled samples with the greatest predictive disparity using Wasserstein Distance, Least Confidence, Entropy Sampling, and Random Sampling. (2) Central line correction: Selected samples are used for expert correction of system-generated tracheal central lines in each training round. (3) Update training dataset: Experts update the training dataset after each DL model's training epoch, enhancing the trustworthiness and performance of the models. (4) Model training: The HCI model is trained using the updated dataset and an enhanced UNet version. Experimental results confirm the effectiveness of these HCI-based approaches, showing that WD-UNet, LC-UNet, UUNet, and RS-UNet achieve comparable or superior performance to state-of-the-art DL models. Notably, WD-UNet achieves this with only 15%-35% of the training data, reducing physician annotation time by 65%-85%.
Diff3Dformer: Leveraging Slice Sequence Diffusion for Enhanced 3D CT Classification with Transformer Networks
Jun 24, 2024Abstract:The manifestation of symptoms associated with lung diseases can vary in different depths for individual patients, highlighting the significance of 3D information in CT scans for medical image classification. While Vision Transformer has shown superior performance over convolutional neural networks in image classification tasks, their effectiveness is often demonstrated on sufficiently large 2D datasets and they easily encounter overfitting issues on small medical image datasets. To address this limitation, we propose a Diffusion-based 3D Vision Transformer (Diff3Dformer), which utilizes the latent space of the Diffusion model to form the slice sequence for 3D analysis and incorporates clustering attention into ViT to aggregate repetitive information within 3D CT scans, thereby harnessing the power of the advanced transformer in 3D classification tasks on small datasets. Our method exhibits improved performance on two different scales of small datasets of 3D lung CT scans, surpassing the state of the art 3D methods and other transformer-based approaches that emerged during the COVID-19 pandemic, demonstrating its robust and superior performance across different scales of data. Experimental results underscore the superiority of our proposed method, indicating its potential for enhancing medical image classification tasks in real-world scenarios.
Fuzzy Attention-based Border Rendering Network for Lung Organ Segmentation
Jun 23, 2024



Abstract:Automatic lung organ segmentation on CT images is crucial for lung disease diagnosis. However, the unlimited voxel values and class imbalance of lung organs can lead to false-negative/positive and leakage issues in advanced methods. Additionally, some slender lung organs are easily lost during the recycled down/up-sample procedure, e.g., bronchioles & arterioles, causing severe discontinuity issue. Inspired by these, this paper introduces an effective lung organ segmentation method called Fuzzy Attention-based Border Rendering (FABR) network. Since fuzzy logic can handle the uncertainty in feature extraction, hence the fusion of deep networks and fuzzy sets should be a viable solution for better performance. Meanwhile, unlike prior top-tier methods that operate on all regular dense points, our FABR depicts lung organ regions as cube-trees, focusing only on recycle-sampled border vulnerable points, rendering the severely discontinuous, false-negative/positive organ regions with a novel Global-Local Cube-tree Fusion (GLCF) module. All experimental results, on four challenging datasets of airway & artery, demonstrate that our method can achieve the favorable performance significantly.
DiffExplainer: Unveiling Black Box Models Via Counterfactual Generation
Jun 21, 2024Abstract:In the field of medical imaging, particularly in tasks related to early disease detection and prognosis, understanding the reasoning behind AI model predictions is imperative for assessing their reliability. Conventional explanation methods encounter challenges in identifying decisive features in medical image classifications, especially when discriminative features are subtle or not immediately evident. To address this limitation, we propose an agent model capable of generating counterfactual images that prompt different decisions when plugged into a black box model. By employing this agent model, we can uncover influential image patterns that impact the black model's final predictions. Through our methodology, we efficiently identify features that influence decisions of the deep black box. We validated our approach in the rigorous domain of medical prognosis tasks, showcasing its efficacy and potential to enhance the reliability of deep learning models in medical image classification compared to existing interpretation methods. The code will be publicly available at https://github.com/ayanglab/DiffExplainer.
When AI Eats Itself: On the Caveats of Data Pollution in the Era of Generative AI
May 15, 2024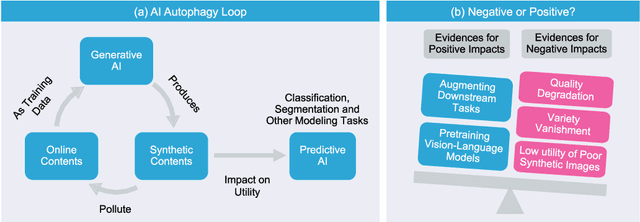
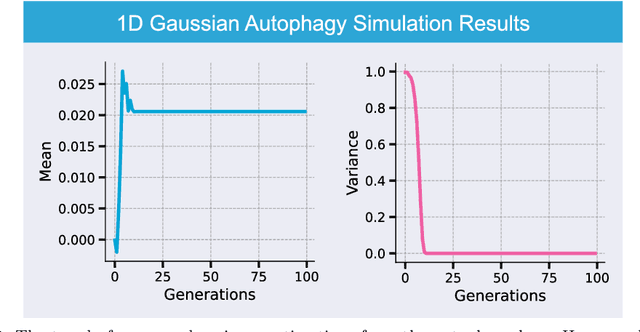
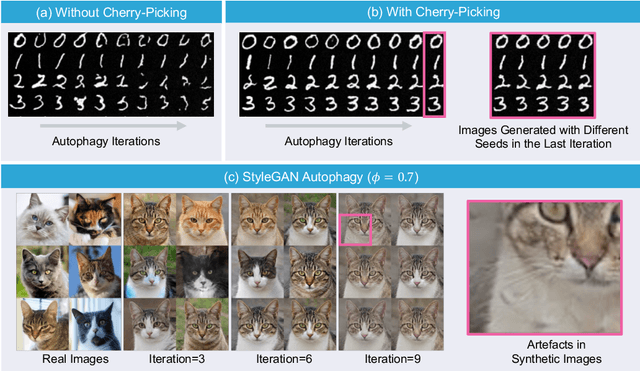

Abstract:Generative artificial intelligence (AI) technologies and large models are producing realistic outputs across various domains, such as images, text, speech, and music. Creating these advanced generative models requires significant resources, particularly large and high-quality datasets. To minimize training expenses, many algorithm developers use data created by the models themselves as a cost-effective training solution. However, not all synthetic data effectively improve model performance, necessitating a strategic balance in the use of real versus synthetic data to optimize outcomes. Currently, the previously well-controlled integration of real and synthetic data is becoming uncontrollable. The widespread and unregulated dissemination of synthetic data online leads to the contamination of datasets traditionally compiled through web scraping, now mixed with unlabeled synthetic data. This trend portends a future where generative AI systems may increasingly rely blindly on consuming self-generated data, raising concerns about model performance and ethical issues. What will happen if generative AI continuously consumes itself without discernment? What measures can we take to mitigate the potential adverse effects? There is a significant gap in the scientific literature regarding the impact of synthetic data use in generative AI, particularly in terms of the fusion of multimodal information. To address this research gap, this review investigates the consequences of integrating synthetic data blindly on training generative AI on both image and text modalities and explores strategies to mitigate these effects. The goal is to offer a comprehensive view of synthetic data's role, advocating for a balanced approach to its use and exploring practices that promote the sustainable development of generative AI technologies in the era of large models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge