Daoqiang Zhang
In-Hospital Stroke Prediction from PPG-Derived Hemodynamic Features
Feb 10, 2026Abstract:The absence of pre-hospital physiological data in standard clinical datasets fundamentally constrains the early prediction of stroke, as patients typically present only after stroke has occurred, leaving the predictive value of continuous monitoring signals such as photoplethysmography (PPG) unvalidated. In this work, we overcome this limitation by focusing on a rare but clinically critical cohort - patients who suffered stroke during hospitalization while already under continuous monitoring - thereby enabling the first large-scale analysis of pre-stroke PPG waveforms aligned to verified onset times. Using MIMIC-III and MC-MED, we develop an LLM-assisted data mining pipeline to extract precise in-hospital stroke onset timestamps from unstructured clinical notes, followed by physician validation, identifying 176 patients (MIMIC) and 158 patients (MC-MED) with high-quality synchronized pre-onset PPG data, respectively. We then extract hemodynamic features from PPG and employ a ResNet-1D model to predict impending stroke across multiple early-warning horizons. The model achieves F1-scores of 0.7956, 0.8759, and 0.9406 at 4, 5, and 6 hours prior to onset on MIMIC-III, and, without re-tuning, reaches 0.9256, 0.9595, and 0.9888 on MC-MED for the same horizons. These results provide the first empirical evidence from real-world clinical data that PPG contains predictive signatures of stroke several hours before onset, demonstrating that passively acquired physiological signals can support reliable early warning, supporting a shift from post-event stroke recognition to proactive, physiology-based surveillance that may materially improve patient outcomes in routine clinical care.
Addressing data annotation scarcity in Brain Tumor Segmentation on 3D MRI scan Using a Semi-Supervised Teacher-Student Framework
Feb 09, 2026Abstract:Accurate brain tumor segmentation from MRI is limited by expensive annotations and data heterogeneity across scanners and sites. We propose a semi-supervised teacher-student framework that combines an uncertainty-aware pseudo-labeling teacher with a progressive, confidence-based curriculum for the student. The teacher produces probabilistic masks and per-pixel uncertainty; unlabeled scans are ranked by image-level confidence and introduced in stages, while a dual-loss objective trains the student to learn from high-confidence regions and unlearn low-confidence ones. Agreement-based refinement further improves pseudo-label quality. On BraTS 2021, validation DSC increased from 0.393 (10% data) to 0.872 (100%), with the largest gains in early stages, demonstrating data efficiency. The teacher reached a validation DSC of 0.922, and the student surpassed the teacher on tumor subregions (e.g., NCR/NET 0.797 and Edema 0.980); notably, the student recovered the Enhancing class (DSC 0.620) where the teacher failed. These results show that confidence-driven curricula and selective unlearning provide robust segmentation under limited supervision and noisy pseudo-labels.
COME: Dual Structure-Semantic Learning with Collaborative MoE for Universal Lesion Detection Across Heterogeneous Ultrasound Datasets
Aug 13, 2025Abstract:Conventional single-dataset training often fails with new data distributions, especially in ultrasound (US) image analysis due to limited data, acoustic shadows, and speckle noise. Therefore, constructing a universal framework for multi-heterogeneous US datasets is imperative. However, a key challenge arises: how to effectively mitigate inter-dataset interference while preserving dataset-specific discriminative features for robust downstream task? Previous approaches utilize either a single source-specific decoder or a domain adaptation strategy, but these methods experienced a decline in performance when applied to other domains. Considering this, we propose a Universal Collaborative Mixture of Heterogeneous Source-Specific Experts (COME). Specifically, COME establishes dual structure-semantic shared experts that create a universal representation space and then collaborate with source-specific experts to extract discriminative features through providing complementary features. This design enables robust generalization by leveraging cross-datasets experience distributions and providing universal US priors for small-batch or unseen data scenarios. Extensive experiments under three evaluation modes (single-dataset, intra-organ, and inter-organ integration datasets) demonstrate COME's superiority, achieving significant mean AP improvements over state-of-the-art methods. Our project is available at: https://universalcome.github.io/UniversalCOME/.
HeLo: Heterogeneous Multi-Modal Fusion with Label Correlation for Emotion Distribution Learning
Jul 09, 2025Abstract:Multi-modal emotion recognition has garnered increasing attention as it plays a significant role in human-computer interaction (HCI) in recent years. Since different discrete emotions may exist at the same time, compared with single-class emotion recognition, emotion distribution learning (EDL) that identifies a mixture of basic emotions has gradually emerged as a trend. However, existing EDL methods face challenges in mining the heterogeneity among multiple modalities. Besides, rich semantic correlations across arbitrary basic emotions are not fully exploited. In this paper, we propose a multi-modal emotion distribution learning framework, named HeLo, aimed at fully exploring the heterogeneity and complementary information in multi-modal emotional data and label correlation within mixed basic emotions. Specifically, we first adopt cross-attention to effectively fuse the physiological data. Then, an optimal transport (OT)-based heterogeneity mining module is devised to mine the interaction and heterogeneity between the physiological and behavioral representations. To facilitate label correlation learning, we introduce a learnable label embedding optimized by correlation matrix alignment. Finally, the learnable label embeddings and label correlation matrices are integrated with the multi-modal representations through a novel label correlation-driven cross-attention mechanism for accurate emotion distribution learning. Experimental results on two publicly available datasets demonstrate the superiority of our proposed method in emotion distribution learning.
MorphSAM: Learning the Morphological Prompts from Atlases for Spine Image Segmentation
Jun 16, 2025Abstract:Spine image segmentation is crucial for clinical diagnosis and treatment of spine diseases. The complex structure of the spine and the high morphological similarity between individual vertebrae and adjacent intervertebral discs make accurate spine segmentation a challenging task. Although the Segment Anything Model (SAM) has been developed, it still struggles to effectively capture and utilize morphological information, limiting its ability to enhance spine image segmentation performance. To address these challenges, in this paper, we propose a MorphSAM that explicitly learns morphological information from atlases, thereby strengthening the spine image segmentation performance of SAM. Specifically, the MorphSAM includes two fully automatic prompt learning networks, 1) an anatomical prompt learning network that directly learns morphological information from anatomical atlases, and 2) a semantic prompt learning network that derives morphological information from text descriptions converted from the atlases. Then, the two learned morphological prompts are fed into the SAM model to boost the segmentation performance. We validate our MorphSAM on two spine image segmentation tasks, including a spine anatomical structure segmentation task with CT images and a lumbosacral plexus segmentation task with MR images. Experimental results demonstrate that our MorphSAM achieves superior segmentation performance when compared to the state-of-the-art methods.
Bridging Distribution Gaps in Time Series Foundation Model Pretraining with Prototype-Guided Normalization
Apr 15, 2025Abstract:Foundation models have achieved remarkable success across diverse machine-learning domains through large-scale pretraining on large, diverse datasets. However, pretraining on such datasets introduces significant challenges due to substantial mismatches in data distributions, a problem particularly pronounced with time series data. In this paper, we tackle this issue by proposing a domain-aware adaptive normalization strategy within the Transformer architecture. Specifically, we replace the traditional LayerNorm with a prototype-guided dynamic normalization mechanism (ProtoNorm), where learned prototypes encapsulate distinct data distributions, and sample-to-prototype affinity determines the appropriate normalization layer. This mechanism effectively captures the heterogeneity of time series characteristics, aligning pretrained representations with downstream tasks. Through comprehensive empirical evaluation, we demonstrate that our method significantly outperforms conventional pretraining techniques across both classification and forecasting tasks, while effectively mitigating the adverse effects of distribution shifts during pretraining. Incorporating ProtoNorm is as simple as replacing a single line of code. Extensive experiments on diverse real-world time series benchmarks validate the robustness and generalizability of our approach, advancing the development of more versatile time series foundation models.
Robust Multimodal Survival Prediction with the Latent Differentiation Conditional Variational AutoEncoder
Mar 12, 2025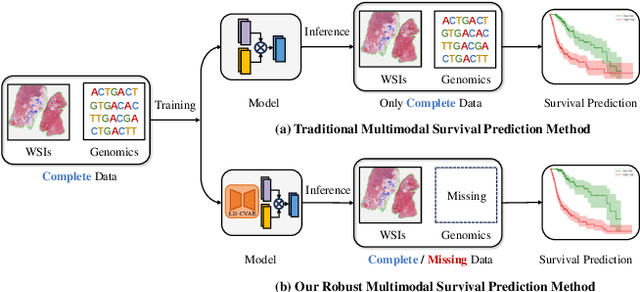

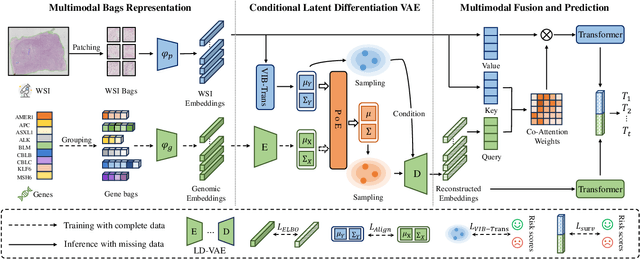

Abstract:The integrative analysis of histopathological images and genomic data has received increasing attention for survival prediction of human cancers. However, the existing studies always hold the assumption that full modalities are available. As a matter of fact, the cost for collecting genomic data is high, which sometimes makes genomic data unavailable in testing samples. A common way of tackling such incompleteness is to generate the genomic representations from the pathology images. Nevertheless, such strategy still faces the following two challenges: (1) The gigapixel whole slide images (WSIs) are huge and thus hard for representation. (2) It is difficult to generate the genomic embeddings with diverse function categories in a unified generative framework. To address the above challenges, we propose a Conditional Latent Differentiation Variational AutoEncoder (LD-CVAE) for robust multimodal survival prediction, even with missing genomic data. Specifically, a Variational Information Bottleneck Transformer (VIB-Trans) module is proposed to learn compressed pathological representations from the gigapixel WSIs. To generate different functional genomic features, we develop a novel Latent Differentiation Variational AutoEncoder (LD-VAE) to learn the common and specific posteriors for the genomic embeddings with diverse functions. Finally, we use the product-of-experts technique to integrate the genomic common posterior and image posterior for the joint latent distribution estimation in LD-CVAE. We test the effectiveness of our method on five different cancer datasets, and the experimental results demonstrate its superiority in both complete and missing modality scenarios.
DAMM-Diffusion: Learning Divergence-Aware Multi-Modal Diffusion Model for Nanoparticles Distribution Prediction
Mar 12, 2025



Abstract:The prediction of nanoparticles (NPs) distribution is crucial for the diagnosis and treatment of tumors. Recent studies indicate that the heterogeneity of tumor microenvironment (TME) highly affects the distribution of NPs across tumors. Hence, it has become a research hotspot to generate the NPs distribution by the aid of multi-modal TME components. However, the distribution divergence among multi-modal TME components may cause side effects i.e., the best uni-modal model may outperform the joint generative model. To address the above issues, we propose a \textbf{D}ivergence-\textbf{A}ware \textbf{M}ulti-\textbf{M}odal \textbf{Diffusion} model (i.e., \textbf{DAMM-Diffusion}) to adaptively generate the prediction results from uni-modal and multi-modal branches in a unified network. In detail, the uni-modal branch is composed of the U-Net architecture while the multi-modal branch extends it by introducing two novel fusion modules i.e., Multi-Modal Fusion Module (MMFM) and Uncertainty-Aware Fusion Module (UAFM). Specifically, the MMFM is proposed to fuse features from multiple modalities, while the UAFM module is introduced to learn the uncertainty map for cross-attention computation. Following the individual prediction results from each branch, the Divergence-Aware Multi-Modal Predictor (DAMMP) module is proposed to assess the consistency of multi-modal data with the uncertainty map, which determines whether the final prediction results come from multi-modal or uni-modal predictions. We predict the NPs distribution given the TME components of tumor vessels and cell nuclei, and the experimental results show that DAMM-Diffusion can generate the distribution of NPs with higher accuracy than the comparing methods. Additional results on the multi-modal brain image synthesis task further validate the effectiveness of the proposed method.
Vision-Language Model IP Protection via Prompt-based Learning
Mar 04, 2025Abstract:Vision-language models (VLMs) like CLIP (Contrastive Language-Image Pre-Training) have seen remarkable success in visual recognition, highlighting the increasing need to safeguard the intellectual property (IP) of well-trained models. Effective IP protection extends beyond ensuring authorized usage; it also necessitates restricting model deployment to authorized data domains, particularly when the model is fine-tuned for specific target domains. However, current IP protection methods often rely solely on the visual backbone, which may lack sufficient semantic richness. To bridge this gap, we introduce IP-CLIP, a lightweight IP protection strategy tailored to CLIP, employing a prompt-based learning approach. By leveraging the frozen visual backbone of CLIP, we extract both image style and content information, incorporating them into the learning of IP prompt. This strategy acts as a robust barrier, effectively preventing the unauthorized transfer of features from authorized domains to unauthorized ones. Additionally, we propose a style-enhancement branch that constructs feature banks for both authorized and unauthorized domains. This branch integrates self-enhanced and cross-domain features, further strengthening IP-CLIP's capability to block features from unauthorized domains. Finally, we present new three metrics designed to better balance the performance degradation of authorized and unauthorized domains. Comprehensive experiments in various scenarios demonstrate its promising potential for application in IP protection tasks for VLMs.
Gaze-Assisted Human-Centric Domain Adaptation for Cardiac Ultrasound Image Segmentation
Feb 06, 2025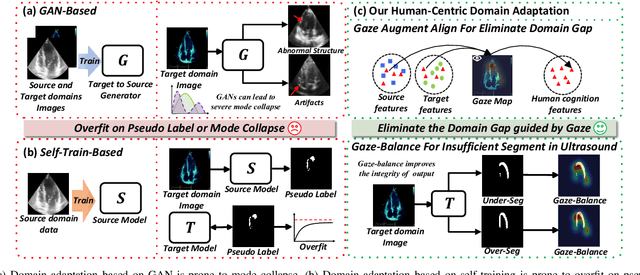
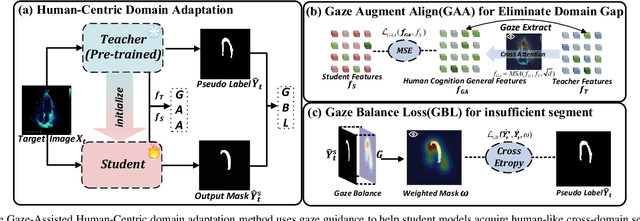
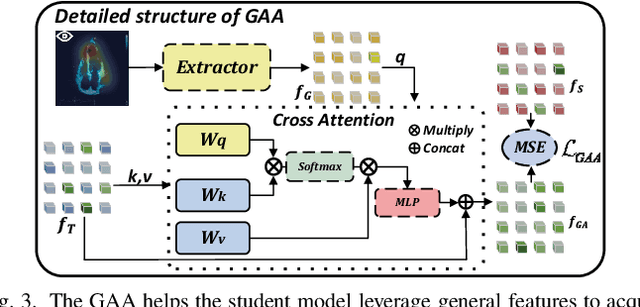
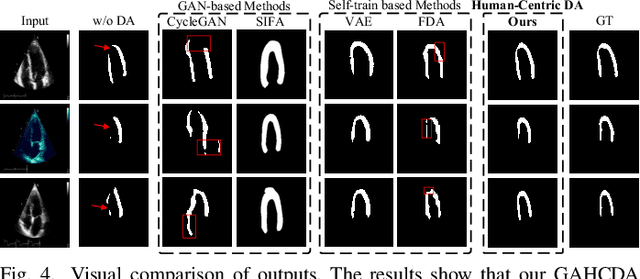
Abstract:Domain adaptation (DA) for cardiac ultrasound image segmentation is clinically significant and valuable. However, previous domain adaptation methods are prone to be affected by the incomplete pseudo-label and low-quality target to source images. Human-centric domain adaptation has great advantages of human cognitive guidance to help model adapt to target domain and reduce reliance on labels. Doctor gaze trajectories contains a large amount of cross-domain human guidance. To leverage gaze information and human cognition for guiding domain adaptation, we propose gaze-assisted human-centric domain adaptation (GAHCDA), which reliably guides the domain adaptation of cardiac ultrasound images. GAHCDA includes following modules: (1) Gaze Augment Alignment (GAA): GAA enables the model to obtain human cognition general features to recognize segmentation target in different domain of cardiac ultrasound images like humans. (2) Gaze Balance Loss (GBL): GBL fused gaze heatmap with outputs which makes the segmentation result structurally closer to the target domain. The experimental results illustrate that our proposed framework is able to segment cardiac ultrasound images more effectively in the target domain than GAN-based methods and other self-train based methods, showing great potential in clinical application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge