Hongen Liao
Native Intelligence Emerges from Large-Scale Clinical Practice: A Retinal Foundation Model with Deployment Efficiency
Dec 16, 2025Abstract:Current retinal foundation models remain constrained by curated research datasets that lack authentic clinical context, and require extensive task-specific optimization for each application, limiting their deployment efficiency in low-resource settings. Here, we show that these barriers can be overcome by building clinical native intelligence directly from real-world medical practice. Our key insight is that large-scale telemedicine programs, where expert centers provide remote consultations across distributed facilities, represent a natural reservoir for learning clinical image interpretation. We present ReVision, a retinal foundation model that learns from the natural alignment between 485,980 color fundus photographs and their corresponding diagnostic reports, accumulated through a decade-long telemedicine program spanning 162 medical institutions across China. Through extensive evaluation across 27 ophthalmic benchmarks, we demonstrate that ReVison enables deployment efficiency with minimal local resources. Without any task-specific training, ReVision achieves zero-shot disease detection with an average AUROC of 0.946 across 12 public benchmarks and 0.952 on 3 independent clinical cohorts. When minimal adaptation is feasible, ReVision matches extensively fine-tuned alternatives while requiring orders of magnitude fewer trainable parameters and labeled examples. The learned representations also transfer effectively to new clinical sites, imaging domains, imaging modalities, and systemic health prediction tasks. In a prospective reader study with 33 ophthalmologists, ReVision's zero-shot assistance improved diagnostic accuracy by 14.8% across all experience levels. These results demonstrate that clinical native intelligence can be directly extracted from clinical archives without any further annotation to build medical AI systems suited to various low-resource settings.
COME: Dual Structure-Semantic Learning with Collaborative MoE for Universal Lesion Detection Across Heterogeneous Ultrasound Datasets
Aug 13, 2025Abstract:Conventional single-dataset training often fails with new data distributions, especially in ultrasound (US) image analysis due to limited data, acoustic shadows, and speckle noise. Therefore, constructing a universal framework for multi-heterogeneous US datasets is imperative. However, a key challenge arises: how to effectively mitigate inter-dataset interference while preserving dataset-specific discriminative features for robust downstream task? Previous approaches utilize either a single source-specific decoder or a domain adaptation strategy, but these methods experienced a decline in performance when applied to other domains. Considering this, we propose a Universal Collaborative Mixture of Heterogeneous Source-Specific Experts (COME). Specifically, COME establishes dual structure-semantic shared experts that create a universal representation space and then collaborate with source-specific experts to extract discriminative features through providing complementary features. This design enables robust generalization by leveraging cross-datasets experience distributions and providing universal US priors for small-batch or unseen data scenarios. Extensive experiments under three evaluation modes (single-dataset, intra-organ, and inter-organ integration datasets) demonstrate COME's superiority, achieving significant mean AP improvements over state-of-the-art methods. Our project is available at: https://universalcome.github.io/UniversalCOME/.
SurGSplat: Progressive Geometry-Constrained Gaussian Splatting for Surgical Scene Reconstruction
Jun 06, 2025Abstract:Intraoperative navigation relies heavily on precise 3D reconstruction to ensure accuracy and safety during surgical procedures. However, endoscopic scenarios present unique challenges, including sparse features and inconsistent lighting, which render many existing Structure-from-Motion (SfM)-based methods inadequate and prone to reconstruction failure. To mitigate these constraints, we propose SurGSplat, a novel paradigm designed to progressively refine 3D Gaussian Splatting (3DGS) through the integration of geometric constraints. By enabling the detailed reconstruction of vascular structures and other critical features, SurGSplat provides surgeons with enhanced visual clarity, facilitating precise intraoperative decision-making. Experimental evaluations demonstrate that SurGSplat achieves superior performance in both novel view synthesis (NVS) and pose estimation accuracy, establishing it as a high-fidelity and efficient solution for surgical scene reconstruction. More information and results can be found on the page https://surgsplat.github.io/.
Hysteresis-Aware Neural Network Modeling and Whole-Body Reinforcement Learning Control of Soft Robots
Apr 18, 2025Abstract:Soft robots exhibit inherent compliance and safety, which makes them particularly suitable for applications requiring direct physical interaction with humans, such as surgical procedures. However, their nonlinear and hysteretic behavior, resulting from the properties of soft materials, presents substantial challenges for accurate modeling and control. In this study, we present a soft robotic system designed for surgical applications and propose a hysteresis-aware whole-body neural network model that accurately captures and predicts the soft robot's whole-body motion, including its hysteretic behavior. Building upon the high-precision dynamic model, we construct a highly parallel simulation environment for soft robot control and apply an on-policy reinforcement learning algorithm to efficiently train whole-body motion control strategies. Based on the trained control policy, we developed a soft robotic system for surgical applications and validated it through phantom-based laser ablation experiments in a physical environment. The results demonstrate that the hysteresis-aware modeling reduces the Mean Squared Error (MSE) by 84.95 percent compared to traditional modeling methods. The deployed control algorithm achieved a trajectory tracking error ranging from 0.126 to 0.250 mm on the real soft robot, highlighting its precision in real-world conditions. The proposed method showed strong performance in phantom-based surgical experiments and demonstrates its potential for complex scenarios, including future real-world clinical applications.
LLM-RG4: Flexible and Factual Radiology Report Generation across Diverse Input Contexts
Dec 16, 2024Abstract:Drafting radiology reports is a complex task requiring flexibility, where radiologists tail content to available information and particular clinical demands. However, most current radiology report generation (RRG) models are constrained to a fixed task paradigm, such as predicting the full ``finding'' section from a single image, inherently involving a mismatch between inputs and outputs. The trained models lack the flexibility for diverse inputs and could generate harmful, input-agnostic hallucinations. To bridge the gap between current RRG models and the clinical demands in practice, we first develop a data generation pipeline to create a new MIMIC-RG4 dataset, which considers four common radiology report drafting scenarios and has perfectly corresponded input and output. Secondly, we propose a novel large language model (LLM) based RRG framework, namely LLM-RG4, which utilizes LLM's flexible instruction-following capabilities and extensive general knowledge. We further develop an adaptive token fusion module that offers flexibility to handle diverse scenarios with different input combinations, while minimizing the additional computational burden associated with increased input volumes. Besides, we propose a token-level loss weighting strategy to direct the model's attention towards positive and uncertain descriptions. Experimental results demonstrate that LLM-RG4 achieves state-of-the-art performance in both clinical efficiency and natural language generation on the MIMIC-RG4 and MIMIC-CXR datasets. We quantitatively demonstrate that our model has minimal input-agnostic hallucinations, whereas current open-source models commonly suffer from this problem.
Anno-incomplete Multi-dataset Detection
Aug 29, 2024



Abstract:Object detectors have shown outstanding performance on various public datasets. However, annotating a new dataset for a new task is usually unavoidable in real, since 1) a single existing dataset usually does not contain all object categories needed; 2) using multiple datasets usually suffers from annotation incompletion and heterogeneous features. We propose a novel problem as "Annotation-incomplete Multi-dataset Detection", and develop an end-to-end multi-task learning architecture which can accurately detect all the object categories with multiple partially annotated datasets. Specifically, we propose an attention feature extractor which helps to mine the relations among different datasets. Besides, a knowledge amalgamation training strategy is incorporated to accommodate heterogeneous features from different sources. Extensive experiments on different object detection datasets demonstrate the effectiveness of our methods and an improvement of 2.17%, 2.10% in mAP can be achieved on COCO and VOC respectively.
Rib Suppression in Digital Chest Tomosynthesis
Mar 05, 2022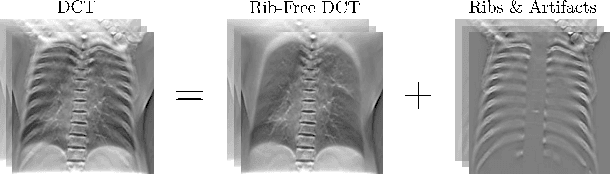
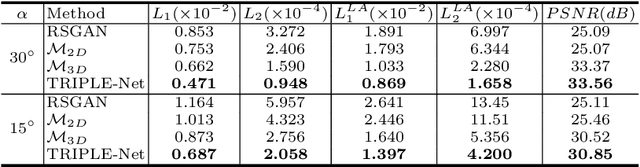
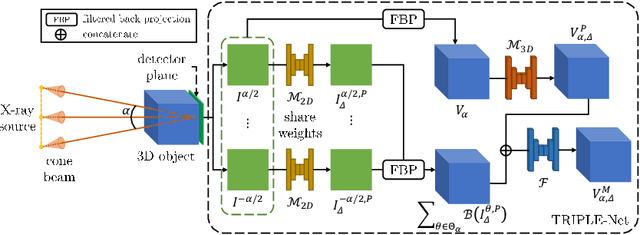
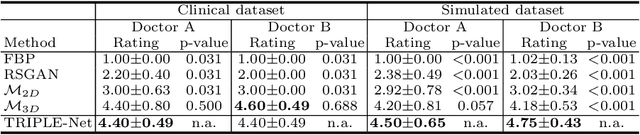
Abstract:Digital chest tomosynthesis (DCT) is a technique to produce sectional 3D images of a human chest for pulmonary disease screening, with 2D X-ray projections taken within an extremely limited range of angles. However, under the limited angle scenario, DCT contains strong artifacts caused by the presence of ribs, jamming the imaging quality of the lung area. Recently, great progress has been achieved for rib suppression in a single X-ray image, to reveal a clearer lung texture. We firstly extend the rib suppression problem to the 3D case at the software level. We propose a $\textbf{T}$omosynthesis $\textbf{RI}$b Su$\textbf{P}$pression and $\textbf{L}$ung $\textbf{E}$nhancement $\textbf{Net}$work (TRIPLE-Net) to model the 3D rib component and provide a rib-free DCT. TRIPLE-Net takes the advantages from both 2D and 3D domains, which model the ribs in DCT with the exact FBP procedure and 3D depth information, respectively. The experiments on simulated datasets and clinical data have shown the effectiveness of TRIPLE-Net to preserve lung details as well as improve the imaging quality of pulmonary diseases. Finally, an expert user study confirms our findings.
A semi-automatic ultrasound image analysis system for the grading diagnosis of COVID-19 pneumonia
Nov 04, 2021


Abstract:This paper proposes a semi-automatic system based on quantitative characterization of the specific image patterns in lung ultrasound (LUS) images, in order to assess the lung conditions of patients with COVID-19 pneumonia, as well as to differentiate between the severe / and no-severe cases. Specifically, four parameters are extracted from each LUS image, namely the thickness (TPL) and roughness (RPL) of the pleural line, and the accumulated with (AWBL) and acoustic coefficient (ACBL) of B lines. 27 patients are enrolled in this study, which are grouped into 13 moderate patients, 7 severe patients and 7 critical patients. Furthermore, the severe and critical patients are regarded as the severe cases, and the moderate patients are regarded as the non-severe cases. Biomarkers among different groups are compared. Each single biomarker and a classifier with all the biomarkers as input are utilized for the binary diagnosis of severe case and non-severe case, respectively. The classifier achieves the best classification performance among all the compared methods (area under the receiver operating characteristics curve = 0.93, sensitivity = 0.93, specificity = 0.85). The proposed image analysis system could be potentially applied to the grading and prognosis evaluation of patients with COVID-19 pneumonia.
Cascaded Feature Warping Network for Unsupervised Medical Image Registration
Mar 15, 2021
Abstract:Deformable image registration is widely utilized in medical image analysis, but most proposed methods fail in the situation of complex deformations. In this paper, we pre-sent a cascaded feature warping network to perform the coarse-to-fine registration. To achieve this, a shared-weights encoder network is adopted to generate the feature pyramids for the unaligned images. The feature warping registration module is then used to estimate the deformation field at each level. The coarse-to-fine manner is implemented by cascading the module from the bottom level to the top level. Furthermore, the multi-scale loss is also introduced to boost the registration performance. We employ two public benchmark datasets and conduct various experiments to evaluate our method. The results show that our method outperforms the state-of-the-art methods, which also demonstrates that the cascaded feature warping network can perform the coarse-to-fine registration effectively and efficiently.
Multi-Modality Generative Adversarial Networks with Tumor Consistency Loss for Brain MR Image Synthesis
May 02, 2020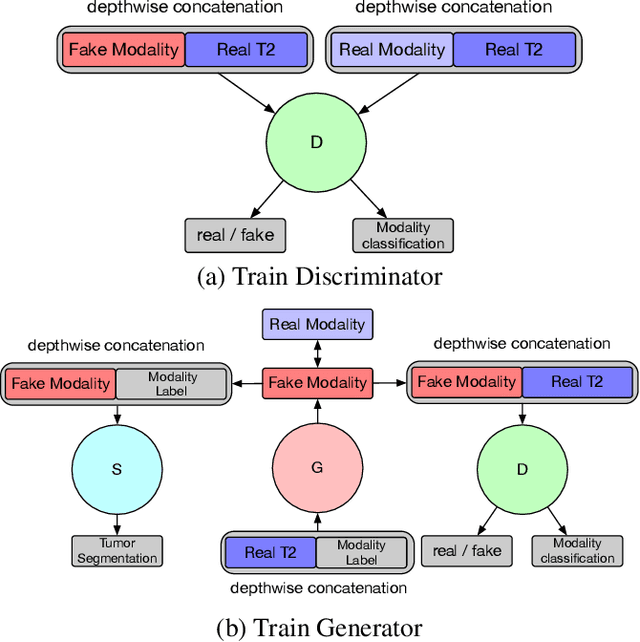
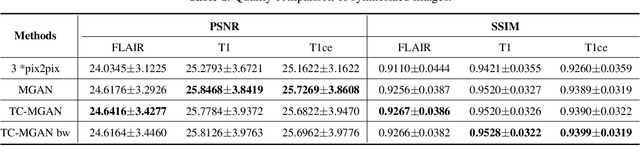
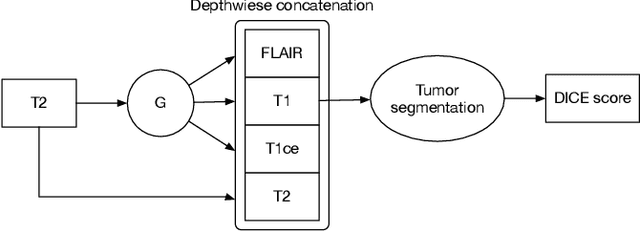
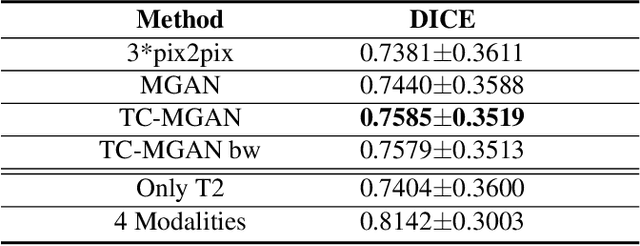
Abstract:Magnetic Resonance (MR) images of different modalities can provide complementary information for clinical diagnosis, but whole modalities are often costly to access. Most existing methods only focus on synthesizing missing images between two modalities, which limits their robustness and efficiency when multiple modalities are missing. To address this problem, we propose a multi-modality generative adversarial network (MGAN) to synthesize three high-quality MR modalities (FLAIR, T1 and T1ce) from one MR modality T2 simultaneously. The experimental results show that the quality of the synthesized images by our proposed methods is better than the one synthesized by the baseline model, pix2pix. Besides, for MR brain image synthesis, it is important to preserve the critical tumor information in the generated modalities, so we further introduce a multi-modality tumor consistency loss to MGAN, called TC-MGAN. We use the synthesized modalities by TC-MGAN to boost the tumor segmentation accuracy, and the results demonstrate its effectiveness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge