Zihan Li
Scale-aware Adaptive Supervised Network with Limited Medical Annotations
Jan 02, 2026Abstract:Medical image segmentation faces critical challenges in semi-supervised learning scenarios due to severe annotation scarcity requiring expert radiological knowledge, significant inter-annotator variability across different viewpoints and expertise levels, and inadequate multi-scale feature integration for precise boundary delineation in complex anatomical structures. Existing semi-supervised methods demonstrate substantial performance degradation compared to fully supervised approaches, particularly in small target segmentation and boundary refinement tasks. To address these fundamental challenges, we propose SASNet (Scale-aware Adaptive Supervised Network), a dual-branch architecture that leverages both low-level and high-level feature representations through novel scale-aware adaptive reweight mechanisms. Our approach introduces three key methodological innovations, including the Scale-aware Adaptive Reweight strategy that dynamically weights pixel-wise predictions using temporal confidence accumulation, the View Variance Enhancement mechanism employing 3D Fourier domain transformations to simulate annotation variability, and segmentation-regression consistency learning through signed distance map algorithms for enhanced boundary precision. These innovations collectively address the core limitations of existing semi-supervised approaches by integrating spatial, temporal, and geometric consistency principles within a unified optimization framework. Comprehensive evaluation across LA, Pancreas-CT, and BraTS datasets demonstrates that SASNet achieves superior performance with limited labeled data, surpassing state-of-the-art semi-supervised methods while approaching fully supervised performance levels. The source code for SASNet is available at https://github.com/HUANGLIZI/SASNet.
Cortex AISQL: A Production SQL Engine for Unstructured Data
Nov 19, 2025Abstract:Snowflake's Cortex AISQL is a production SQL engine that integrates native semantic operations directly into SQL. This integration allows users to write declarative queries that combine relational operations with semantic reasoning, enabling them to query both structured and unstructured data effortlessly. However, making semantic operations efficient at production scale poses fundamental challenges. Semantic operations are more expensive than traditional SQL operations, possess distinct latency and throughput characteristics, and their cost and selectivity are unknown during query compilation. Furthermore, existing query engines are not designed to optimize semantic operations. The AISQL query execution engine addresses these challenges through three novel techniques informed by production deployment data from Snowflake customers. First, AI-aware query optimization treats AI inference cost as a first-class optimization objective, reasoning about large language model (LLM) cost directly during query planning to achieve 2-8$\times$ speedups. Second, adaptive model cascades reduce inference costs by routing most rows through a fast proxy model while escalating uncertain cases to a powerful oracle model, achieving 2-6$\times$ speedups while maintaining 90-95% of oracle model quality. Third, semantic join query rewriting lowers the quadratic time complexity of join operations to linear through reformulation as multi-label classification tasks, achieving 15-70$\times$ speedups with often improved prediction quality. AISQL is deployed in production at Snowflake, where it powers diverse customer workloads across analytics, search, and content understanding.
SF-Recon: Simplification-Free Lightweight Building Reconstruction via 3D Gaussian Splatting
Nov 17, 2025Abstract:Lightweight building surface models are crucial for digital city, navigation, and fast geospatial analytics, yet conventional multi-view geometry pipelines remain cumbersome and quality-sensitive due to their reliance on dense reconstruction, meshing, and subsequent simplification. This work presents SF-Recon, a method that directly reconstructs lightweight building surfaces from multi-view images without post-hoc mesh simplification. We first train an initial 3D Gaussian Splatting (3DGS) field to obtain a view-consistent representation. Building structure is then distilled by a normal-gradient-guided Gaussian optimization that selects primitives aligned with roof and wall boundaries, followed by multi-view edge-consistency pruning to enhance structural sharpness and suppress non-structural artifacts without external supervision. Finally, a multi-view depth-constrained Delaunay triangulation converts the structured Gaussian field into a lightweight, structurally faithful building mesh. Based on a proposed SF dataset, the experimental results demonstrate that our SF-Recon can directly reconstruct lightweight building models from multi-view imagery, achieving substantially fewer faces and vertices while maintaining computational efficiency. Website:https://lzh282140127-cell.github.io/SF-Recon-project/
DETECT: Data-Driven Evaluation of Treatments Enabled by Classification Transformers
Nov 10, 2025Abstract:Chronic pain is a global health challenge affecting millions of individuals, making it essential for physicians to have reliable and objective methods to measure the functional impact of clinical treatments. Traditionally used methods, like the numeric rating scale, while personalized and easy to use, are subjective due to their self-reported nature. Thus, this paper proposes DETECT (Data-Driven Evaluation of Treatments Enabled by Classification Transformers), a data-driven framework that assesses treatment success by comparing patient activities of daily life before and after treatment. We use DETECT on public benchmark datasets and simulated patient data from smartphone sensors. Our results demonstrate that DETECT is objective yet lightweight, making it a significant and novel contribution to clinical decision-making. By using DETECT, independently or together with other self-reported metrics, physicians can improve their understanding of their treatment impacts, ultimately leading to more personalized and responsive patient care.
TCM-Eval: An Expert-Level Dynamic and Extensible Benchmark for Traditional Chinese Medicine
Nov 10, 2025Abstract:Large Language Models (LLMs) have demonstrated remarkable capabilities in modern medicine, yet their application in Traditional Chinese Medicine (TCM) remains severely limited by the absence of standardized benchmarks and the scarcity of high-quality training data. To address these challenges, we introduce TCM-Eval, the first dynamic and extensible benchmark for TCM, meticulously curated from national medical licensing examinations and validated by TCM experts. Furthermore, we construct a large-scale training corpus and propose Self-Iterative Chain-of-Thought Enhancement (SI-CoTE) to autonomously enrich question-answer pairs with validated reasoning chains through rejection sampling, establishing a virtuous cycle of data and model co-evolution. Using this enriched training data, we develop ZhiMingTang (ZMT), a state-of-the-art LLM specifically designed for TCM, which significantly exceeds the passing threshold for human practitioners. To encourage future research and development, we release a public leaderboard, fostering community engagement and continuous improvement.
RFI Detection and Identification at OVRO Using Pseudonymetry
Oct 31, 2025



Abstract:Protecting radio astronomy observatories from unintended interference is critical as wireless transmissions increases near protected bands. While database-driven coordination frameworks and radio quiet zones exist, they cannot rapidly identify or suppress specific interfering transmitters, especially at low signal-to-noise ratio (SNR) levels. This paper presents the first over-the-air field demonstration of Pseudonymetry at the Owens Valley Radio Observatory (OVRO), illustrating cooperative spectrum sharing between heterogeneous wireless systems. In our experiment, a narrow-band secondary transmitter embeds a pseudonym watermark into its signal, while the wide-band radio telescope passively extracts the watermark from spectrogram data. Results show that interference can be reliably detected and the interfering device uniquely identified even at low SNR where conventional demodulation is infeasible. These findings validate that passive scientific receivers can participate in a lightweight feedback loop to trigger shutdown of harmful transmissions, demonstrating the potential of Pseudonymetry as a complementary enforcement tool for protecting radio astronomy environments.
Sensing and Stopping Interfering Secondary Users: Validation of an Efficient Spectrum Sharing System
Jul 17, 2025Abstract:We present the design and validation of Stoppable Secondary Use (StopSec), a privacy-preserving protocol with the capability to identify a secondary user (SU) causing interference to a primary user (PU) and to act quickly to stop the interference. All users are served by a database that provides a feedback mechanism from a PU to an interfering SU. We introduce a new lightweight and robust method to watermark an SU's OFDM packet. Through extensive over-the-air real-time experiments, we evaluate StopSec in terms of interference detection, identification, and stopping latency, as well as impact on SUs. We show that the watermarking method avoids negative impact to the secondary data link and is robust to real-world time-varying channels. Interfering SUs can be stopped in under 150 milliseconds, and when multiple users are simultaneously interfering, they can all be stopped. Even when the interference is 10 dB lower than the noise power, StopSec successfully stops interfering SUs within a few seconds of their appearance in the channel. StopSec can be an effective spectrum sharing protocol for cases when interference to a PU must be quickly and automatically stopped.
Kling-Foley: Multimodal Diffusion Transformer for High-Quality Video-to-Audio Generation
Jun 24, 2025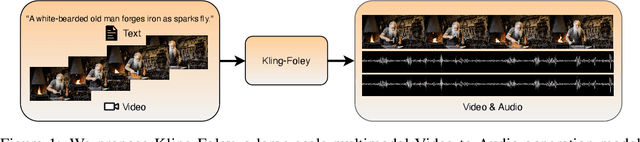
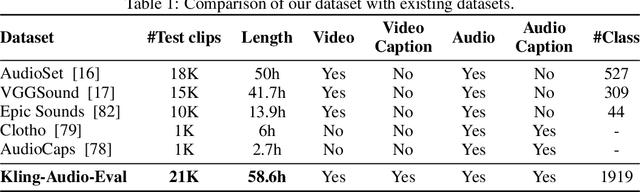
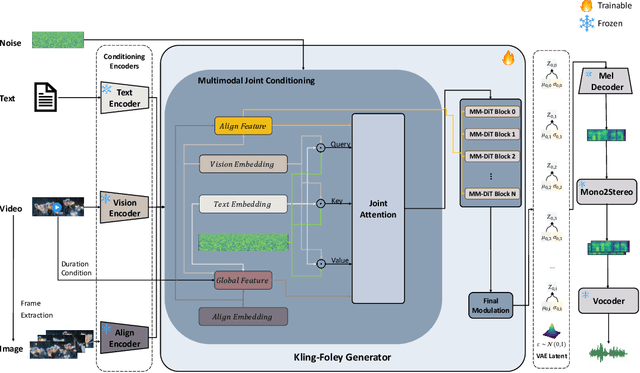
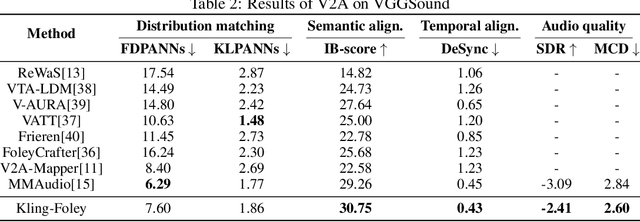
Abstract:We propose Kling-Foley, a large-scale multimodal Video-to-Audio generation model that synthesizes high-quality audio synchronized with video content. In Kling-Foley, we introduce multimodal diffusion transformers to model the interactions between video, audio, and text modalities, and combine it with a visual semantic representation module and an audio-visual synchronization module to enhance alignment capabilities. Specifically, these modules align video conditions with latent audio elements at the frame level, thereby improving semantic alignment and audio-visual synchronization. Together with text conditions, this integrated approach enables precise generation of video-matching sound effects. In addition, we propose a universal latent audio codec that can achieve high-quality modeling in various scenarios such as sound effects, speech, singing, and music. We employ a stereo rendering method that imbues synthesized audio with a spatial presence. At the same time, in order to make up for the incomplete types and annotations of the open-source benchmark, we also open-source an industrial-level benchmark Kling-Audio-Eval. Our experiments show that Kling-Foley trained with the flow matching objective achieves new audio-visual SOTA performance among public models in terms of distribution matching, semantic alignment, temporal alignment and audio quality.
FIC-TSC: Learning Time Series Classification with Fisher Information Constraint
May 09, 2025



Abstract:Analyzing time series data is crucial to a wide spectrum of applications, including economics, online marketplaces, and human healthcare. In particular, time series classification plays an indispensable role in segmenting different phases in stock markets, predicting customer behavior, and classifying worker actions and engagement levels. These aspects contribute significantly to the advancement of automated decision-making and system optimization in real-world applications. However, there is a large consensus that time series data often suffers from domain shifts between training and test sets, which dramatically degrades the classification performance. Despite the success of (reversible) instance normalization in handling the domain shifts for time series regression tasks, its performance in classification is unsatisfactory. In this paper, we propose \textit{FIC-TSC}, a training framework for time series classification that leverages Fisher information as the constraint. We theoretically and empirically show this is an efficient and effective solution to guide the model converge toward flatter minima, which enhances its generalizability to distribution shifts. We rigorously evaluate our method on 30 UEA multivariate and 85 UCR univariate datasets. Our empirical results demonstrate the superiority of the proposed method over 14 recent state-of-the-art methods.
STPNet: Scale-aware Text Prompt Network for Medical Image Segmentation
Apr 02, 2025Abstract:Accurate segmentation of lesions plays a critical role in medical image analysis and diagnosis. Traditional segmentation approaches that rely solely on visual features often struggle with the inherent uncertainty in lesion distribution and size. To address these issues, we propose STPNet, a Scale-aware Text Prompt Network that leverages vision-language modeling to enhance medical image segmentation. Our approach utilizes multi-scale textual descriptions to guide lesion localization and employs retrieval-segmentation joint learning to bridge the semantic gap between visual and linguistic modalities. Crucially, STPNet retrieves relevant textual information from a specialized medical text repository during training, eliminating the need for text input during inference while retaining the benefits of cross-modal learning. We evaluate STPNet on three datasets: COVID-Xray, COVID-CT, and Kvasir-SEG. Experimental results show that our vision-language approach outperforms state-of-the-art segmentation methods, demonstrating the effectiveness of incorporating textual semantic knowledge into medical image analysis. The code has been made publicly on https://github.com/HUANGLIZI/STPNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge