Xiangde Luo
University of Electronic Science and Technology of China, Chengdu, China, ShangAI Laboratory, Shanghai, China
A3-TTA: Adaptive Anchor Alignment Test-Time Adaptation for Image Segmentation
Feb 03, 2026Abstract:Test-Time Adaptation (TTA) offers a practical solution for deploying image segmentation models under domain shift without accessing source data or retraining. Among existing TTA strategies, pseudo-label-based methods have shown promising performance. However, they often rely on perturbation-ensemble heuristics (e.g., dropout sampling, test-time augmentation, Gaussian noise), which lack distributional grounding and yield unstable training signals. This can trigger error accumulation and catastrophic forgetting during adaptation. To address this, we propose \textbf{A3-TTA}, a TTA framework that constructs reliable pseudo-labels through anchor-guided supervision. Specifically, we identify well-predicted target domain images using a class compact density metric, under the assumption that confident predictions imply distributional proximity to the source domain. These anchors serve as stable references to guide pseudo-label generation, which is further regularized via semantic consistency and boundary-aware entropy minimization. Additionally, we introduce a self-adaptive exponential moving average strategy to mitigate label noise and stabilize model update during adaptation. Evaluated on both multi-domain medical images (heart structure and prostate segmentation) and natural images, A3-TTA significantly improves average Dice scores by 10.40 to 17.68 percentage points compared to the source model, outperforming several state-of-the-art TTA methods under different segmentation model architectures. A3-TTA also excels in continual TTA, maintaining high performance across sequential target domains with strong anti-forgetting ability. The code will be made publicly available at https://github.com/HiLab-git/A3-TTA.
nnMIL: A generalizable multiple instance learning framework for computational pathology
Nov 18, 2025Abstract:Computational pathology holds substantial promise for improving diagnosis and guiding treatment decisions. Recent pathology foundation models enable the extraction of rich patch-level representations from large-scale whole-slide images (WSIs), but current approaches for aggregating these features into slide-level predictions remain constrained by design limitations that hinder generalizability and reliability. Here, we developed nnMIL, a simple yet broadly applicable multiple-instance learning framework that connects patch-level foundation models to robust slide-level clinical inference. nnMIL introduces random sampling at both the patch and feature levels, enabling large-batch optimization, task-aware sampling strategies, and efficient and scalable training across datasets and model architectures. A lightweight aggregator performs sliding-window inference to generate ensemble slide-level predictions and supports principled uncertainty estimation. Across 40,000 WSIs encompassing 35 clinical tasks and four pathology foundation models, nnMIL consistently outperformed existing MIL methods for disease diagnosis, histologic subtyping, molecular biomarker detection, and pan- cancer prognosis prediction. It further demonstrated strong cross-model generalization, reliable uncertainty quantification, and robust survival stratification in multiple external cohorts. In conclusion, nnMIL offers a practical and generalizable solution for translating pathology foundation models into clinically meaningful predictions, advancing the development and deployment of reliable AI systems in real-world settings.
A Generative Foundation Model for Chest Radiography
Sep 04, 2025Abstract:The scarcity of well-annotated diverse medical images is a major hurdle for developing reliable AI models in healthcare. Substantial technical advances have been made in generative foundation models for natural images. Here we develop `ChexGen', a generative vision-language foundation model that introduces a unified framework for text-, mask-, and bounding box-guided synthesis of chest radiographs. Built upon the latent diffusion transformer architecture, ChexGen was pretrained on the largest curated chest X-ray dataset to date, consisting of 960,000 radiograph-report pairs. ChexGen achieves accurate synthesis of radiographs through expert evaluations and quantitative metrics. We demonstrate the utility of ChexGen for training data augmentation and supervised pretraining, which led to performance improvements across disease classification, detection, and segmentation tasks using a small fraction of training data. Further, our model enables the creation of diverse patient cohorts that enhance model fairness by detecting and mitigating demographic biases. Our study supports the transformative role of generative foundation models in building more accurate, data-efficient, and equitable medical AI systems.
DiffOSeg: Omni Medical Image Segmentation via Multi-Expert Collaboration Diffusion Model
Jul 17, 2025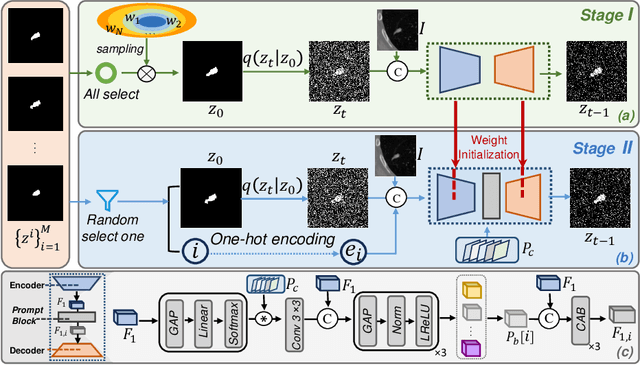
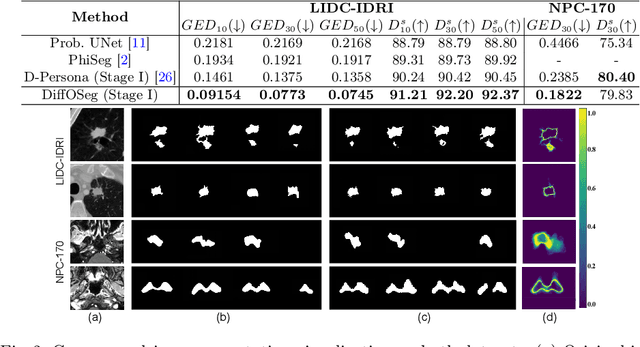
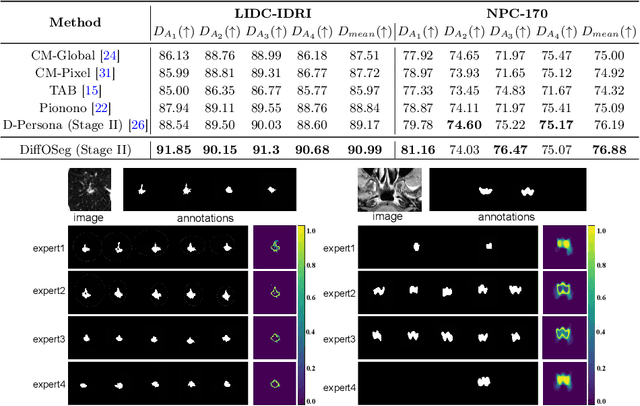

Abstract:Annotation variability remains a substantial challenge in medical image segmentation, stemming from ambiguous imaging boundaries and diverse clinical expertise. Traditional deep learning methods producing single deterministic segmentation predictions often fail to capture these annotator biases. Although recent studies have explored multi-rater segmentation, existing methods typically focus on a single perspective -- either generating a probabilistic ``gold standard'' consensus or preserving expert-specific preferences -- thus struggling to provide a more omni view. In this study, we propose DiffOSeg, a two-stage diffusion-based framework, which aims to simultaneously achieve both consensus-driven (combining all experts' opinions) and preference-driven (reflecting experts' individual assessments) segmentation. Stage I establishes population consensus through a probabilistic consensus strategy, while Stage II captures expert-specific preference via adaptive prompts. Demonstrated on two public datasets (LIDC-IDRI and NPC-170), our model outperforms existing state-of-the-art methods across all evaluated metrics. Source code is available at https://github.com/string-ellipses/DiffOSeg .
Cycle-Consistent Bridge Diffusion Model for Accelerated MRI Reconstruction
Dec 13, 2024



Abstract:Accelerated MRI reconstruction techniques aim to reduce examination time while maintaining high image fidelity, which is highly desirable in clinical settings for improving patient comfort and hospital efficiency. Existing deep learning methods typically reconstruct images from under-sampled data with traditional reconstruction approaches, but they still struggle to provide high-fidelity results. Diffusion models show great potential to improve fidelity of generated images in recent years. However, their inference process starting with a random Gaussian noise introduces instability into the results and usually requires thousands of sampling steps, resulting in sub-optimal reconstruction quality and low efficiency. To address these challenges, we propose Cycle-Consistent Bridge Diffusion Model (CBDM). CBDM employs two bridge diffusion models to construct a cycle-consistent diffusion process with a consistency loss, enhancing the fine-grained details of reconstructed images and reducing the number of diffusion steps. Moreover, CBDM incorporates a Contourlet Decomposition Embedding Module (CDEM) which captures multi-scale structural texture knowledge in images through frequency domain decomposition pyramids and directional filter banks to improve structural fidelity. Extensive experiments demonstrate the superiority of our model by higher reconstruction quality and fewer training iterations, achieving a new state of the art for accelerated MRI reconstruction in both fastMRI and IXI datasets.
Cross Group Attention and Group-wise Rolling for Multimodal Medical Image Synthesis
Nov 22, 2024Abstract:Multimodal MR image synthesis aims to generate missing modality image by fusing and mapping a few available MRI data. Most existing approaches typically adopt an image-to-image translation scheme. However, these methods often suffer from sub-optimal performance due to the spatial misalignment between different modalities while they are typically treated as input channels. Therefore, in this paper, we propose an Adaptive Group-wise Interaction Network (AGI-Net) that explores both inter-modality and intra-modality relationships for multimodal MR image synthesis. Specifically, groups are first pre-defined along the channel dimension and then we perform an adaptive rolling for the standard convolutional kernel to capture inter-modality spatial correspondences. At the same time, a cross-group attention module is introduced to fuse information across different channel groups, leading to better feature representation. We evaluated the effectiveness of our model on the publicly available IXI and BraTS2023 datasets, where the AGI-Net achieved state-of-the-art performance for multimodal MR image synthesis. Code will be released.
Weakly Supervised Lymph Nodes Segmentation Based on Partial Instance Annotations with Pre-trained Dual-branch Network and Pseudo Label Learning
Aug 18, 2024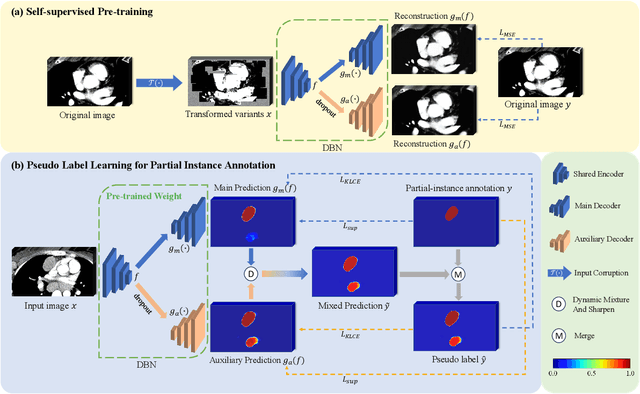
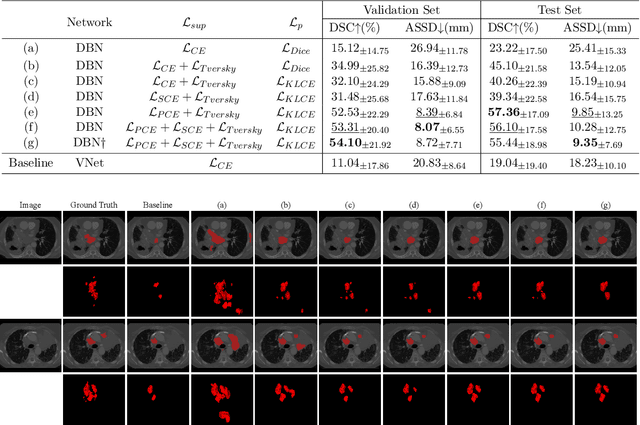
Abstract:Assessing the presence of potentially malignant lymph nodes aids in estimating cancer progression, and identifying surrounding benign lymph nodes can assist in determining potential metastatic pathways for cancer. For quantitative analysis, automatic segmentation of lymph nodes is crucial. However, due to the labor-intensive and time-consuming manual annotation process required for a large number of lymph nodes, it is more practical to annotate only a subset of the lymph node instances to reduce annotation costs. In this study, we propose a pre-trained Dual-Branch network with Dynamically Mixed Pseudo label (DBDMP) to learn from partial instance annotations for lymph nodes segmentation. To obtain reliable pseudo labels for lymph nodes that are not annotated, we employ a dual-decoder network to generate different outputs that are then dynamically mixed. We integrate the original weak partial annotations with the mixed pseudo labels to supervise the network. To further leverage the extensive amount of unannotated voxels, we apply a self-supervised pre-training strategy to enhance the model's feature extraction capability. Experiments on the mediastinal Lymph Node Quantification (LNQ) dataset demonstrate that our method, compared to directly learning from partial instance annotations, significantly improves the Dice Similarity Coefficient (DSC) from 11.04% to 54.10% and reduces the Average Symmetric Surface Distance (ASSD) from 20.83 $mm$ to 8.72 $mm$. The code is available at https://github.com/WltyBY/LNQ2023_training_code.git
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:013
SAM-Driven Weakly Supervised Nodule Segmentation with Uncertainty-Aware Cross Teaching
Jul 18, 2024



Abstract:Automated nodule segmentation is essential for computer-assisted diagnosis in ultrasound images. Nevertheless, most existing methods depend on precise pixel-level annotations by medical professionals, a process that is both costly and labor-intensive. Recently, segmentation foundation models like SAM have shown impressive generalizability on natural images, suggesting their potential as pseudo-labelers. However, accurate prompts remain crucial for their success in medical images. In this work, we devise a novel weakly supervised framework that effectively utilizes the segmentation foundation model to generate pseudo-labels from aspect ration annotations for automatic nodule segmentation. Specifically, we develop three types of bounding box prompts based on scalable shape priors, followed by an adaptive pseudo-label selection module to fully exploit the prediction capabilities of the foundation model for nodules. We also present a SAM-driven uncertainty-aware cross-teaching strategy. This approach integrates SAM-based uncertainty estimation and label-space perturbations into cross-teaching to mitigate the impact of pseudo-label inaccuracies on model training. Extensive experiments on two clinically collected ultrasound datasets demonstrate the superior performance of our proposed method.
An Uncertainty-guided Tiered Self-training Framework for Active Source-free Domain Adaptation in Prostate Segmentation
Jul 03, 2024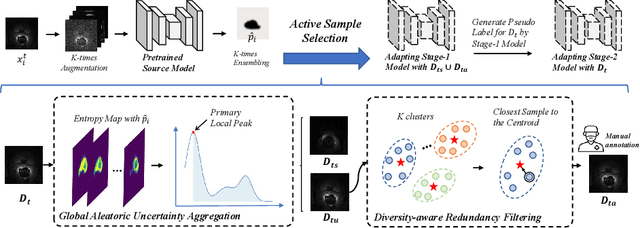
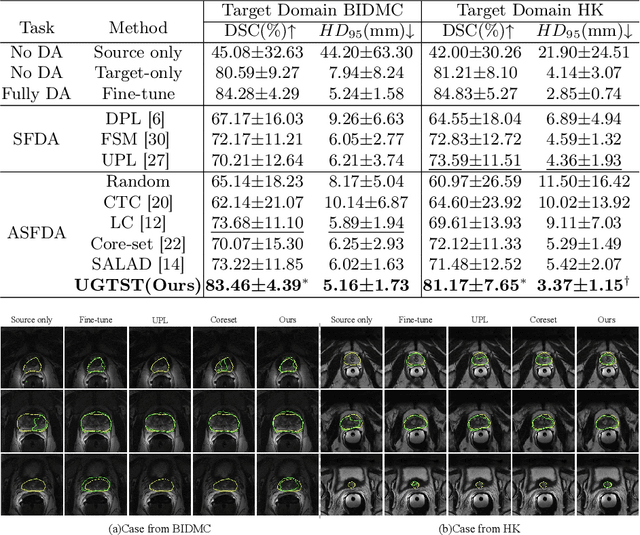
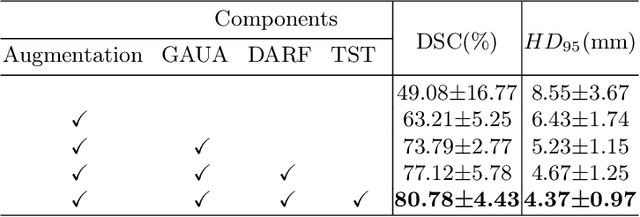
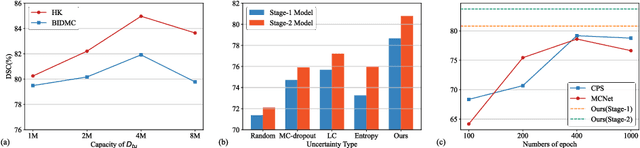
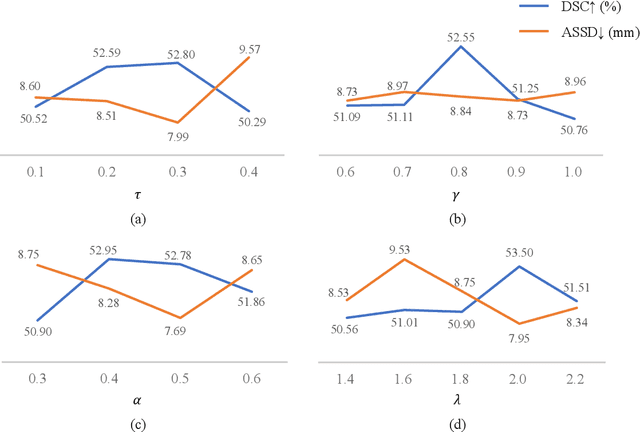
Abstract:Deep learning models have exhibited remarkable efficacy in accurately delineating the prostate for diagnosis and treatment of prostate diseases, but challenges persist in achieving robust generalization across different medical centers. Source-free Domain Adaptation (SFDA) is a promising technique to adapt deep segmentation models to address privacy and security concerns while reducing domain shifts between source and target domains. However, recent literature indicates that the performance of SFDA remains far from satisfactory due to unpredictable domain gaps. Annotating a few target domain samples is acceptable, as it can lead to significant performance improvement with a low annotation cost. Nevertheless, due to extremely limited annotation budgets, careful consideration is needed in selecting samples for annotation. Inspired by this, our goal is to develop Active Source-free Domain Adaptation (ASFDA) for medical image segmentation. Specifically, we propose a novel Uncertainty-guided Tiered Self-training (UGTST) framework, consisting of efficient active sample selection via entropy-based primary local peak filtering to aggregate global uncertainty and diversity-aware redundancy filter, coupled with a tiered self-learning strategy, achieves stable domain adaptation. Experimental results on cross-center prostate MRI segmentation datasets revealed that our method yielded marked advancements, with a mere 5% annotation, exhibiting an average Dice score enhancement of 9.78% and 7.58% in two target domains compared with state-of-the-art methods, on par with fully supervised learning. Code is available at:https://github.com/HiLab-git/UGTST
Advancing UWF-SLO Vessel Segmentation with Source-Free Active Domain Adaptation and a Novel Multi-Center Dataset
Jun 19, 2024Abstract:Accurate vessel segmentation in Ultra-Wide-Field Scanning Laser Ophthalmoscopy (UWF-SLO) images is crucial for diagnosing retinal diseases. Although recent techniques have shown encouraging outcomes in vessel segmentation, models trained on one medical dataset often underperform on others due to domain shifts. Meanwhile, manually labeling high-resolution UWF-SLO images is an extremely challenging, time-consuming and expensive task. In response, this study introduces a pioneering framework that leverages a patch-based active domain adaptation approach. By actively recommending a few valuable image patches by the devised Cascade Uncertainty-Predominance (CUP) selection strategy for labeling and model-finetuning, our method significantly improves the accuracy of UWF-SLO vessel segmentation across diverse medical centers. In addition, we annotate and construct the first Multi-center UWF-SLO Vessel Segmentation (MU-VS) dataset to promote this topic research, comprising data from multiple institutions. This dataset serves as a valuable resource for cross-center evaluation, verifying the effectiveness and robustness of our approach. Experimental results demonstrate that our approach surpasses existing domain adaptation and active learning methods, considerably reducing the gap between the Upper and Lower bounds with minimal annotations, highlighting our method's practical clinical value. We will release our dataset and code to facilitate relevant research: https://github.com/whq-xxh/SFADA-UWF-SLO.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge