Litingyu Wang
University of Electronic Science and Technology of China, Chengdu, China
SegRap2025: A Benchmark of Gross Tumor Volume and Lymph Node Clinical Target Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Jan 28, 2026Abstract:Accurate delineation of Gross Tumor Volume (GTV), Lymph Node Clinical Target Volume (LN CTV), and Organ-at-Risk (OAR) from Computed Tomography (CT) scans is essential for precise radiotherapy planning in Nasopharyngeal Carcinoma (NPC). Building upon SegRap2023, which focused on OAR and GTV segmentation using single-center paired non-contrast CT (ncCT) and contrast-enhanced CT (ceCT) scans, the SegRap2025 challenge aims to enhance the generalizability and robustness of segmentation models across imaging centers and modalities. SegRap2025 comprises two tasks: Task01 addresses GTV segmentation using paired CT from the SegRap2023 dataset, with an additional external testing set to evaluate cross-center generalization, and Task02 focuses on LN CTV segmentation using multi-center training data and an unseen external testing set, where each case contains paired CT scans or a single modality, emphasizing both cross-center and cross-modality robustness. This paper presents the challenge setup and provides a comprehensive analysis of the solutions submitted by ten participating teams. For GTV segmentation task, the top-performing models achieved average Dice Similarity Coefficient (DSC) of 74.61% and 56.79% on the internal and external testing cohorts, respectively. For LN CTV segmentation task, the highest average DSC values reached 60.24%, 60.50%, and 57.23% on paired CT, ceCT-only, and ncCT-only subsets, respectively. SegRap2025 establishes a large-scale multi-center, multi-modality benchmark for evaluating the generalization and robustness in radiotherapy target segmentation, providing valuable insights toward clinically applicable automated radiotherapy planning systems. The benchmark is available at: https://hilab-git.github.io/SegRap2025_Challenge.
Head and Neck Tumor Segmentation of MRI from Pre- and Mid-radiotherapy with Pre-training, Data Augmentation and Dual Flow UNet
Dec 19, 2024



Abstract:Head and neck tumors and metastatic lymph nodes are crucial for treatment planning and prognostic analysis. Accurate segmentation and quantitative analysis of these structures require pixel-level annotation, making automated segmentation techniques essential for the diagnosis and treatment of head and neck cancer. In this study, we investigated the effects of multiple strategies on the segmentation of pre-radiotherapy (pre-RT) and mid-radiotherapy (mid-RT) images. For the segmentation of pre-RT images, we utilized: 1) a fully supervised learning approach, and 2) the same approach enhanced with pre-trained weights and the MixUp data augmentation technique. For mid-RT images, we introduced a novel computational-friendly network architecture that features separate encoders for mid-RT images and registered pre-RT images with their labels. The mid-RT encoder branch integrates information from pre-RT images and labels progressively during the forward propagation. We selected the highest-performing model from each fold and used their predictions to create an ensemble average for inference. In the final test, our models achieved a segmentation performance of 82.38% for pre-RT and 72.53% for mid-RT on aggregated Dice Similarity Coefficient (DSC) as HiLab. Our code is available at https://github.com/WltyBY/HNTS-MRG2024_train_code.
LNQ 2023 challenge: Benchmark of weakly-supervised techniques for mediastinal lymph node quantification
Aug 19, 2024



Abstract:Accurate assessment of lymph node size in 3D CT scans is crucial for cancer staging, therapeutic management, and monitoring treatment response. Existing state-of-the-art segmentation frameworks in medical imaging often rely on fully annotated datasets. However, for lymph node segmentation, these datasets are typically small due to the extensive time and expertise required to annotate the numerous lymph nodes in 3D CT scans. Weakly-supervised learning, which leverages incomplete or noisy annotations, has recently gained interest in the medical imaging community as a potential solution. Despite the variety of weakly-supervised techniques proposed, most have been validated only on private datasets or small publicly available datasets. To address this limitation, the Mediastinal Lymph Node Quantification (LNQ) challenge was organized in conjunction with the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023). This challenge aimed to advance weakly-supervised segmentation methods by providing a new, partially annotated dataset and a robust evaluation framework. A total of 16 teams from 5 countries submitted predictions to the validation leaderboard, and 6 teams from 3 countries participated in the evaluation phase. The results highlighted both the potential and the current limitations of weakly-supervised approaches. On one hand, weakly-supervised approaches obtained relatively good performance with a median Dice score of $61.0\%$. On the other hand, top-ranked teams, with a median Dice score exceeding $70\%$, boosted their performance by leveraging smaller but fully annotated datasets to combine weak supervision and full supervision. This highlights both the promise of weakly-supervised methods and the ongoing need for high-quality, fully annotated data to achieve higher segmentation performance.
Weakly Supervised Lymph Nodes Segmentation Based on Partial Instance Annotations with Pre-trained Dual-branch Network and Pseudo Label Learning
Aug 18, 2024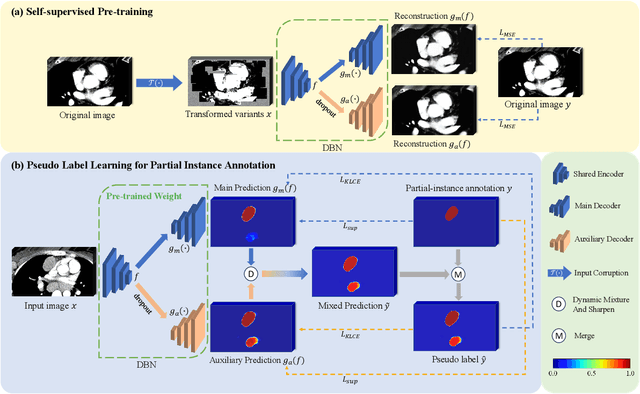
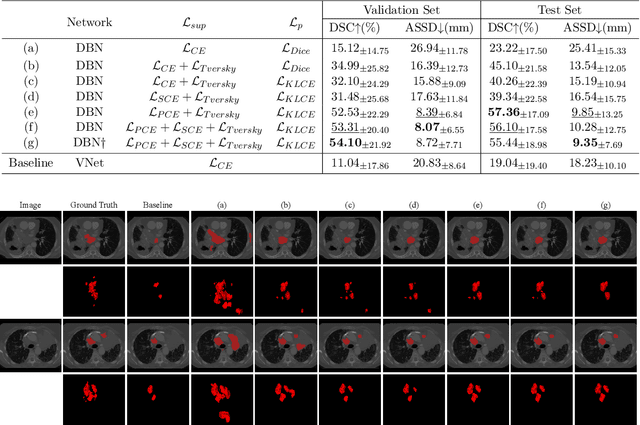
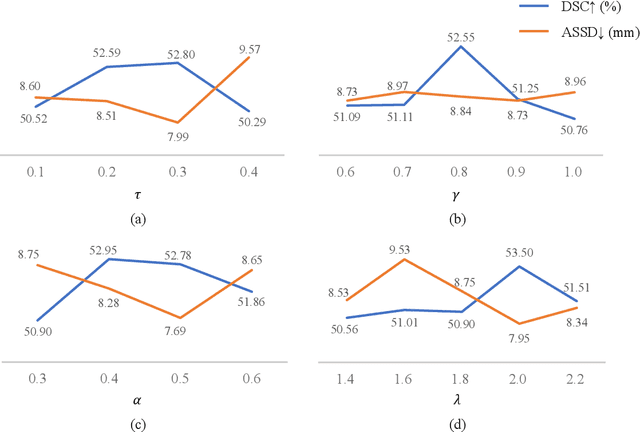
Abstract:Assessing the presence of potentially malignant lymph nodes aids in estimating cancer progression, and identifying surrounding benign lymph nodes can assist in determining potential metastatic pathways for cancer. For quantitative analysis, automatic segmentation of lymph nodes is crucial. However, due to the labor-intensive and time-consuming manual annotation process required for a large number of lymph nodes, it is more practical to annotate only a subset of the lymph node instances to reduce annotation costs. In this study, we propose a pre-trained Dual-Branch network with Dynamically Mixed Pseudo label (DBDMP) to learn from partial instance annotations for lymph nodes segmentation. To obtain reliable pseudo labels for lymph nodes that are not annotated, we employ a dual-decoder network to generate different outputs that are then dynamically mixed. We integrate the original weak partial annotations with the mixed pseudo labels to supervise the network. To further leverage the extensive amount of unannotated voxels, we apply a self-supervised pre-training strategy to enhance the model's feature extraction capability. Experiments on the mediastinal Lymph Node Quantification (LNQ) dataset demonstrate that our method, compared to directly learning from partial instance annotations, significantly improves the Dice Similarity Coefficient (DSC) from 11.04% to 54.10% and reduces the Average Symmetric Surface Distance (ASSD) from 20.83 $mm$ to 8.72 $mm$. The code is available at https://github.com/WltyBY/LNQ2023_training_code.git
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:013
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge