Shichuan Zhang
University of Electronic Science and Technology of China, Chengdu, China, Department of Radiation Oncology, Sichuan Cancer Hospital & Institute, Sichuan Cancer Center, Chengdu, China
SegRap2025: A Benchmark of Gross Tumor Volume and Lymph Node Clinical Target Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Jan 28, 2026Abstract:Accurate delineation of Gross Tumor Volume (GTV), Lymph Node Clinical Target Volume (LN CTV), and Organ-at-Risk (OAR) from Computed Tomography (CT) scans is essential for precise radiotherapy planning in Nasopharyngeal Carcinoma (NPC). Building upon SegRap2023, which focused on OAR and GTV segmentation using single-center paired non-contrast CT (ncCT) and contrast-enhanced CT (ceCT) scans, the SegRap2025 challenge aims to enhance the generalizability and robustness of segmentation models across imaging centers and modalities. SegRap2025 comprises two tasks: Task01 addresses GTV segmentation using paired CT from the SegRap2023 dataset, with an additional external testing set to evaluate cross-center generalization, and Task02 focuses on LN CTV segmentation using multi-center training data and an unseen external testing set, where each case contains paired CT scans or a single modality, emphasizing both cross-center and cross-modality robustness. This paper presents the challenge setup and provides a comprehensive analysis of the solutions submitted by ten participating teams. For GTV segmentation task, the top-performing models achieved average Dice Similarity Coefficient (DSC) of 74.61% and 56.79% on the internal and external testing cohorts, respectively. For LN CTV segmentation task, the highest average DSC values reached 60.24%, 60.50%, and 57.23% on paired CT, ceCT-only, and ncCT-only subsets, respectively. SegRap2025 establishes a large-scale multi-center, multi-modality benchmark for evaluating the generalization and robustness in radiotherapy target segmentation, providing valuable insights toward clinically applicable automated radiotherapy planning systems. The benchmark is available at: https://hilab-git.github.io/SegRap2025_Challenge.
OpenPath: Open-Set Active Learning for Pathology Image Classification via Pre-trained Vision-Language Models
Jun 18, 2025Abstract:Pathology image classification plays a crucial role in accurate medical diagnosis and treatment planning. Training high-performance models for this task typically requires large-scale annotated datasets, which are both expensive and time-consuming to acquire. Active Learning (AL) offers a solution by iteratively selecting the most informative samples for annotation, thereby reducing the labeling effort. However, most AL methods are designed under the assumption of a closed-set scenario, where all the unannotated images belong to target classes. In real-world clinical environments, the unlabeled pool often contains a substantial amount of Out-Of-Distribution (OOD) data, leading to low efficiency of annotation in traditional AL methods. Furthermore, most existing AL methods start with random selection in the first query round, leading to a significant waste of labeling costs in open-set scenarios. To address these challenges, we propose OpenPath, a novel open-set active learning approach for pathological image classification leveraging a pre-trained Vision-Language Model (VLM). In the first query, we propose task-specific prompts that combine target and relevant non-target class prompts to effectively select In-Distribution (ID) and informative samples from the unlabeled pool. In subsequent queries, Diverse Informative ID Sampling (DIS) that includes Prototype-based ID candidate Selection (PIS) and Entropy-Guided Stochastic Sampling (EGSS) is proposed to ensure both purity and informativeness in a query, avoiding the selection of OOD samples. Experiments on two public pathology image datasets show that OpenPath significantly enhances the model's performance due to its high purity of selected samples, and outperforms several state-of-the-art open-set AL methods. The code is available at \href{https://github.com/HiLab-git/OpenPath}{https://github.com/HiLab-git/OpenPath}..
CLISC: Bridging clip and sam by enhanced cam for unsupervised brain tumor segmentation
Jan 27, 2025



Abstract:Brain tumor segmentation is important for diagnosis of the tumor, and current deep-learning methods rely on a large set of annotated images for training, with high annotation costs. Unsupervised segmentation is promising to avoid human annotations while the performance is often limited. In this study, we present a novel unsupervised segmentation approach that leverages the capabilities of foundation models, and it consists of three main steps: (1) A vision-language model (i.e., CLIP) is employed to obtain image-level pseudo-labels for training a classification network. Class Activation Mapping (CAM) is then employed to extract Regions of Interest (ROIs), where an adaptive masking-based data augmentation is used to enhance ROI identification.(2) The ROIs are used to generate bounding box and point prompts for the Segment Anything Model (SAM) to obtain segmentation pseudo-labels. (3) A 3D segmentation network is trained with the SAM-derived pseudo-labels, where low-quality pseudo-labels are filtered out in a self-learning process based on the similarity between the SAM's output and the network's prediction. Evaluation on the BraTS2020 dataset demonstrates that our approach obtained an average Dice Similarity Score (DSC) of 85.60%, outperforming five state-of-the-art unsupervised segmentation methods by more than 10 percentage points. Besides, our approach outperforms directly using SAM for zero-shot inference, and its performance is close to fully supervised learning.
Head and Neck Tumor Segmentation of MRI from Pre- and Mid-radiotherapy with Pre-training, Data Augmentation and Dual Flow UNet
Dec 19, 2024



Abstract:Head and neck tumors and metastatic lymph nodes are crucial for treatment planning and prognostic analysis. Accurate segmentation and quantitative analysis of these structures require pixel-level annotation, making automated segmentation techniques essential for the diagnosis and treatment of head and neck cancer. In this study, we investigated the effects of multiple strategies on the segmentation of pre-radiotherapy (pre-RT) and mid-radiotherapy (mid-RT) images. For the segmentation of pre-RT images, we utilized: 1) a fully supervised learning approach, and 2) the same approach enhanced with pre-trained weights and the MixUp data augmentation technique. For mid-RT images, we introduced a novel computational-friendly network architecture that features separate encoders for mid-RT images and registered pre-RT images with their labels. The mid-RT encoder branch integrates information from pre-RT images and labels progressively during the forward propagation. We selected the highest-performing model from each fold and used their predictions to create an ensemble average for inference. In the final test, our models achieved a segmentation performance of 82.38% for pre-RT and 72.53% for mid-RT on aggregated Dice Similarity Coefficient (DSC) as HiLab. Our code is available at https://github.com/WltyBY/HNTS-MRG2024_train_code.
Weakly Supervised Lymph Nodes Segmentation Based on Partial Instance Annotations with Pre-trained Dual-branch Network and Pseudo Label Learning
Aug 18, 2024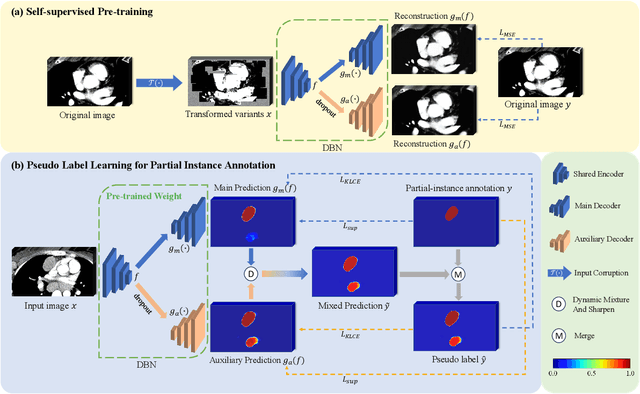
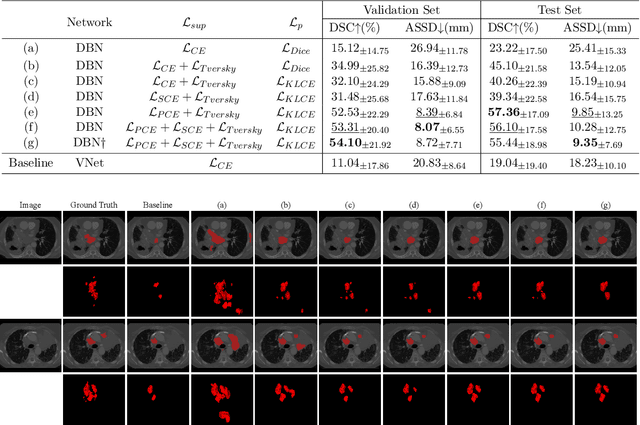
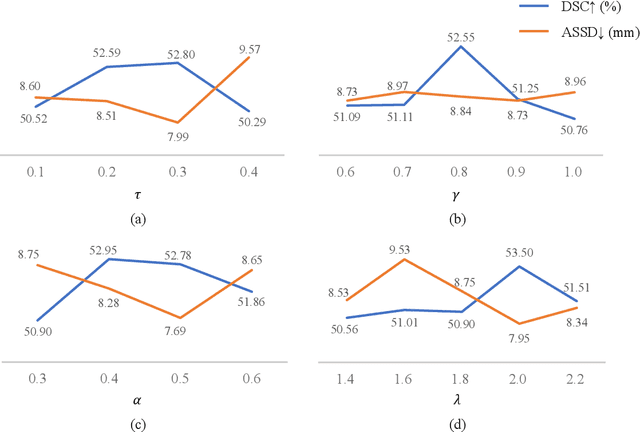
Abstract:Assessing the presence of potentially malignant lymph nodes aids in estimating cancer progression, and identifying surrounding benign lymph nodes can assist in determining potential metastatic pathways for cancer. For quantitative analysis, automatic segmentation of lymph nodes is crucial. However, due to the labor-intensive and time-consuming manual annotation process required for a large number of lymph nodes, it is more practical to annotate only a subset of the lymph node instances to reduce annotation costs. In this study, we propose a pre-trained Dual-Branch network with Dynamically Mixed Pseudo label (DBDMP) to learn from partial instance annotations for lymph nodes segmentation. To obtain reliable pseudo labels for lymph nodes that are not annotated, we employ a dual-decoder network to generate different outputs that are then dynamically mixed. We integrate the original weak partial annotations with the mixed pseudo labels to supervise the network. To further leverage the extensive amount of unannotated voxels, we apply a self-supervised pre-training strategy to enhance the model's feature extraction capability. Experiments on the mediastinal Lymph Node Quantification (LNQ) dataset demonstrate that our method, compared to directly learning from partial instance annotations, significantly improves the Dice Similarity Coefficient (DSC) from 11.04% to 54.10% and reduces the Average Symmetric Surface Distance (ASSD) from 20.83 $mm$ to 8.72 $mm$. The code is available at https://github.com/WltyBY/LNQ2023_training_code.git
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:013
Large-scale cervical precancerous screening via AI-assisted cytology whole slide image analysis
Jul 28, 2024



Abstract:Cervical Cancer continues to be the leading gynecological malignancy, posing a persistent threat to women's health on a global scale. Early screening via cytology Whole Slide Image (WSI) diagnosis is critical to prevent this Cancer progression and improve survival rate, but pathologist's single test suffers inevitable false negative due to the immense number of cells that need to be reviewed within a WSI. Though computer-aided automated diagnostic models can serve as strong complement for pathologists, their effectiveness is hampered by the paucity of extensive and detailed annotations, coupled with the limited interpretability and robustness. These factors significantly hinder their practical applicability and reliability in clinical settings. To tackle these challenges, we develop an AI approach, which is a Scalable Technology for Robust and Interpretable Diagnosis built on Extensive data (STRIDE) of cervical cytology. STRIDE addresses the bottleneck of limited annotations by integrating patient-level labels with a small portion of cell-level labels through an end-to-end training strategy, facilitating scalable learning across extensive datasets. To further improve the robustness to real-world domain shifts of cytology slide-making and imaging, STRIDE employs color adversarial samples training that mimic staining and imaging variations. Lastly, to achieve pathologist-level interpretability for the trustworthiness in clinical settings, STRIDE can generate explanatory textual descriptions that simulates pathologists' diagnostic processes by cell image feature and textual description alignment. Conducting extensive experiments and evaluations in 183 medical centers with a dataset of 341,889 WSIs and 0.1 billion cells from cervical cytology patients, STRIDE has demonstrated a remarkable superiority over previous state-of-the-art techniques.
Combining Supervised Learning and Reinforcement Learning for Multi-Label Classification Tasks with Partial Labels
Jun 24, 2024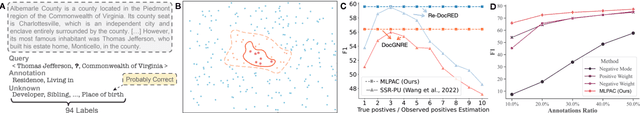
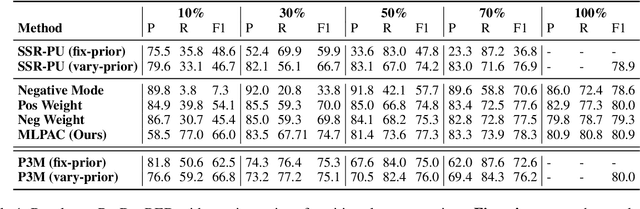
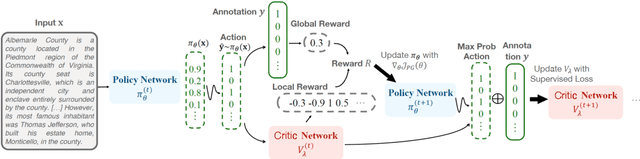
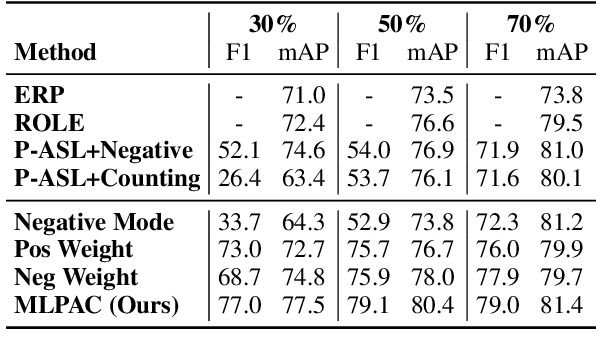
Abstract:Traditional supervised learning heavily relies on human-annotated datasets, especially in data-hungry neural approaches. However, various tasks, especially multi-label tasks like document-level relation extraction, pose challenges in fully manual annotation due to the specific domain knowledge and large class sets. Therefore, we address the multi-label positive-unlabelled learning (MLPUL) problem, where only a subset of positive classes is annotated. We propose Mixture Learner for Partially Annotated Classification (MLPAC), an RL-based framework combining the exploration ability of reinforcement learning and the exploitation ability of supervised learning. Experimental results across various tasks, including document-level relation extraction, multi-label image classification, and binary PU learning, demonstrate the generalization and effectiveness of our framework.
Multi-modal Learning with Missing Modality in Predicting Axillary Lymph Node Metastasis
Jan 03, 2024



Abstract:Multi-modal Learning has attracted widespread attention in medical image analysis. Using multi-modal data, whole slide images (WSIs) and clinical information, can improve the performance of deep learning models in the diagnosis of axillary lymph node metastasis. However, clinical information is not easy to collect in clinical practice due to privacy concerns, limited resources, lack of interoperability, etc. Although patient selection can ensure the training set to have multi-modal data for model development, missing modality of clinical information can appear during test. This normally leads to performance degradation, which limits the use of multi-modal models in the clinic. To alleviate this problem, we propose a bidirectional distillation framework consisting of a multi-modal branch and a single-modal branch. The single-modal branch acquires the complete multi-modal knowledge from the multi-modal branch, while the multi-modal learns the robust features of WSI from the single-modal. We conduct experiments on a public dataset of Lymph Node Metastasis in Early Breast Cancer to validate the method. Our approach not only achieves state-of-the-art performance with an AUC of 0.861 on the test set without missing data, but also yields an AUC of 0.842 when the rate of missing modality is 80\%. This shows the effectiveness of the approach in dealing with multi-modal data and missing modality. Such a model has the potential to improve treatment decision-making for early breast cancer patients who have axillary lymph node metastatic status.
SegRap2023: A Benchmark of Organs-at-Risk and Gross Tumor Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Dec 15, 2023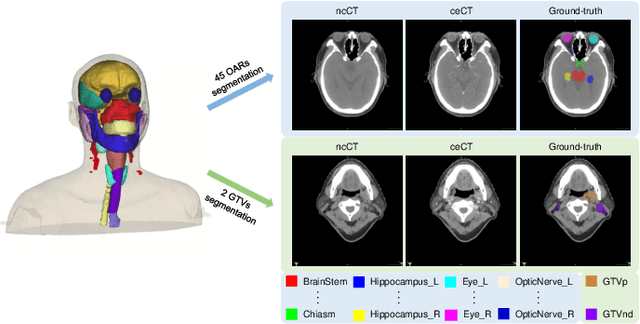
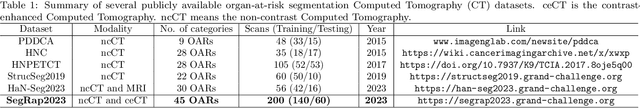
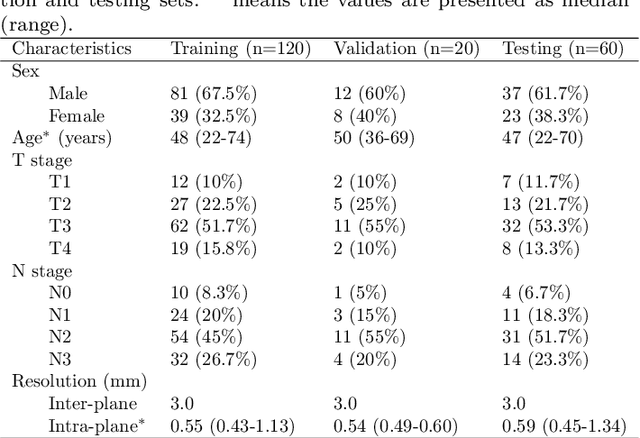
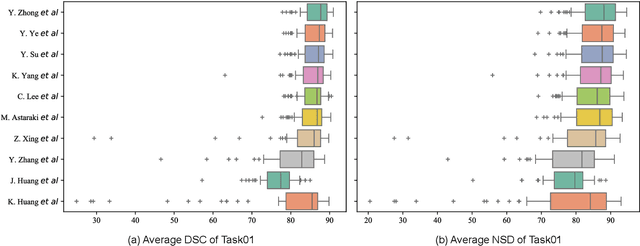
Abstract:Radiation therapy is a primary and effective NasoPharyngeal Carcinoma (NPC) treatment strategy. The precise delineation of Gross Tumor Volumes (GTVs) and Organs-At-Risk (OARs) is crucial in radiation treatment, directly impacting patient prognosis. Previously, the delineation of GTVs and OARs was performed by experienced radiation oncologists. Recently, deep learning has achieved promising results in many medical image segmentation tasks. However, for NPC OARs and GTVs segmentation, few public datasets are available for model development and evaluation. To alleviate this problem, the SegRap2023 challenge was organized in conjunction with MICCAI2023 and presented a large-scale benchmark for OAR and GTV segmentation with 400 Computed Tomography (CT) scans from 200 NPC patients, each with a pair of pre-aligned non-contrast and contrast-enhanced CT scans. The challenge's goal was to segment 45 OARs and 2 GTVs from the paired CT scans. In this paper, we detail the challenge and analyze the solutions of all participants. The average Dice similarity coefficient scores for all submissions ranged from 76.68\% to 86.70\%, and 70.42\% to 73.44\% for OARs and GTVs, respectively. We conclude that the segmentation of large-size OARs is well-addressed, and more efforts are needed for GTVs and small-size or thin-structure OARs. The benchmark will remain publicly available here: https://segrap2023.grand-challenge.org
Scribble-based 3D Multiple Abdominal Organ Segmentation via Triple-branch Multi-dilated Network with Pixel- and Class-wise Consistency
Sep 18, 2023

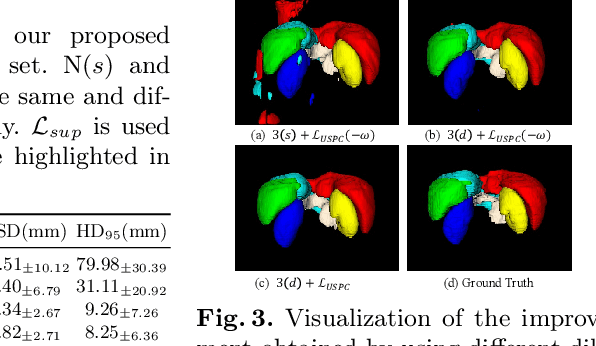
Abstract:Multi-organ segmentation in abdominal Computed Tomography (CT) images is of great importance for diagnosis of abdominal lesions and subsequent treatment planning. Though deep learning based methods have attained high performance, they rely heavily on large-scale pixel-level annotations that are time-consuming and labor-intensive to obtain. Due to its low dependency on annotation, weakly supervised segmentation has attracted great attention. However, there is still a large performance gap between current weakly-supervised methods and fully supervised learning, leaving room for exploration. In this work, we propose a novel 3D framework with two consistency constraints for scribble-supervised multiple abdominal organ segmentation from CT. Specifically, we employ a Triple-branch multi-Dilated network (TDNet) with one encoder and three decoders using different dilation rates to capture features from different receptive fields that are complementary to each other to generate high-quality soft pseudo labels. For more stable unsupervised learning, we use voxel-wise uncertainty to rectify the soft pseudo labels and then supervise the outputs of each decoder. To further regularize the network, class relationship information is exploited by encouraging the generated class affinity matrices to be consistent across different decoders under multi-view projection. Experiments on the public WORD dataset show that our method outperforms five existing scribble-supervised methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge