Pingyi Chen
Towards Effective and Efficient Context-aware Nucleus Detection in Histopathology Whole Slide Images
Mar 04, 2025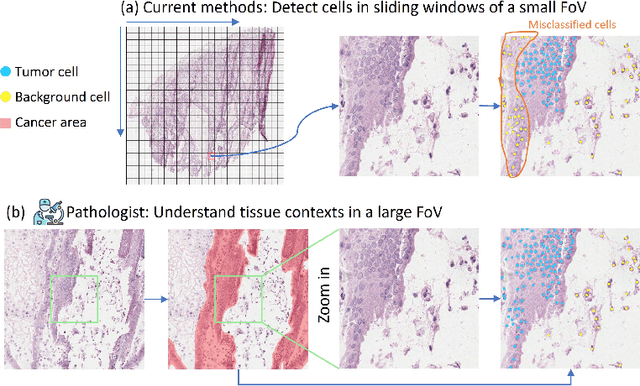
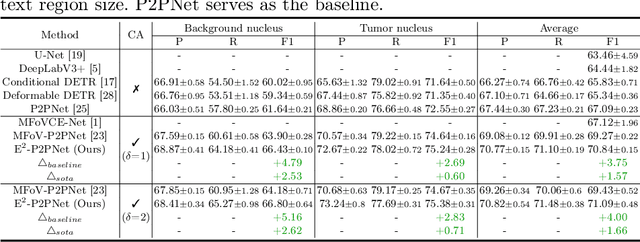
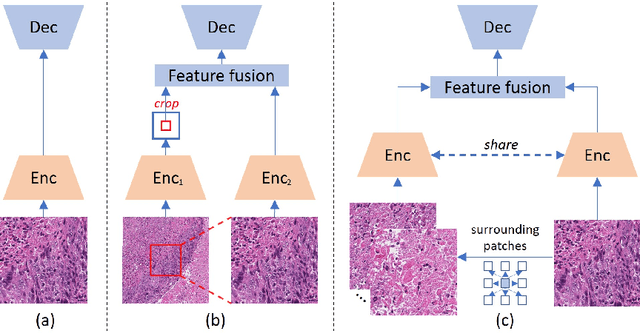
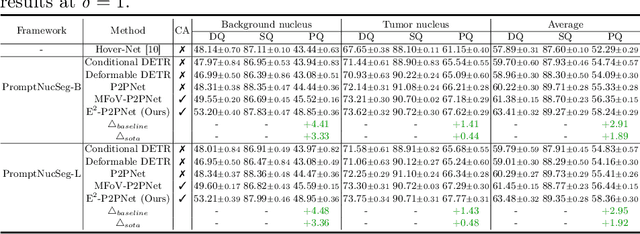
Abstract:Nucleus detection in histopathology whole slide images (WSIs) is crucial for a broad spectrum of clinical applications. Current approaches for nucleus detection in gigapixel WSIs utilize a sliding window methodology, which overlooks boarder contextual information (eg, tissue structure) and easily leads to inaccurate predictions. To address this problem, recent studies additionally crops a large Filed-of-View (FoV) region around each sliding window to extract contextual features. However, such methods substantially increases the inference latency. In this paper, we propose an effective and efficient context-aware nucleus detection algorithm. Specifically, instead of leveraging large FoV regions, we aggregate contextual clues from off-the-shelf features of historically visited sliding windows. This design greatly reduces computational overhead. Moreover, compared to large FoV regions at a low magnification, the sliding window patches have higher magnification and provide finer-grained tissue details, thereby enhancing the detection accuracy. To further improve the efficiency, we propose a grid pooling technique to compress dense feature maps of each patch into a few contextual tokens. Finally, we craft OCELOT-seg, the first benchmark dedicated to context-aware nucleus instance segmentation. Code, dataset, and model checkpoints will be available at https://github.com/windygoo/PathContext.
CPath-Omni: A Unified Multimodal Foundation Model for Patch and Whole Slide Image Analysis in Computational Pathology
Dec 16, 2024Abstract:The emergence of large multimodal models (LMMs) has brought significant advancements to pathology. Previous research has primarily focused on separately training patch-level and whole-slide image (WSI)-level models, limiting the integration of learned knowledge across patches and WSIs, and resulting in redundant models. In this work, we introduce CPath-Omni, the first 15-billion-parameter LMM designed to unify both patch and WSI level image analysis, consolidating a variety of tasks at both levels, including classification, visual question answering, captioning, and visual referring prompting. Extensive experiments demonstrate that CPath-Omni achieves state-of-the-art (SOTA) performance across seven diverse tasks on 39 out of 42 datasets, outperforming or matching task-specific models trained for individual tasks. Additionally, we develop a specialized pathology CLIP-based visual processor for CPath-Omni, CPath-CLIP, which, for the first time, integrates different vision models and incorporates a large language model as a text encoder to build a more powerful CLIP model, which achieves SOTA performance on nine zero-shot and four few-shot datasets. Our findings highlight CPath-Omni's ability to unify diverse pathology tasks, demonstrating its potential to streamline and advance the field of foundation model in pathology.
Rethinking Transformer for Long Contextual Histopathology Whole Slide Image Analysis
Oct 18, 2024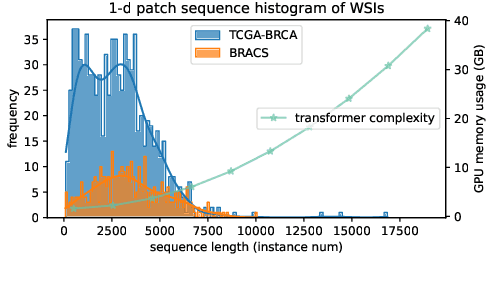
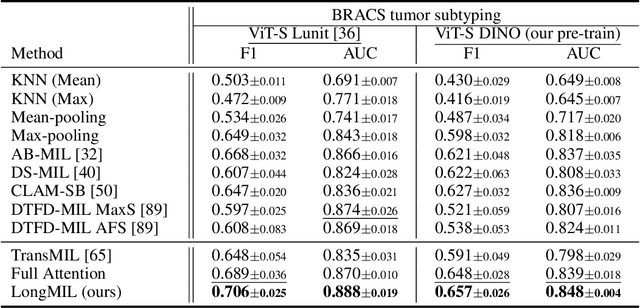
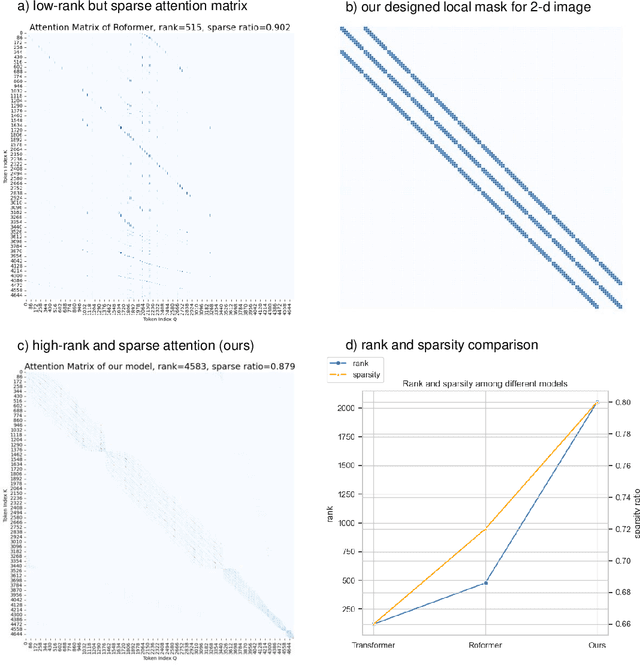
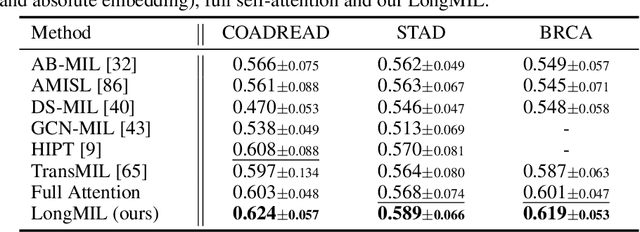
Abstract:Histopathology Whole Slide Image (WSI) analysis serves as the gold standard for clinical cancer diagnosis in the daily routines of doctors. To develop computer-aided diagnosis model for WSIs, previous methods typically employ Multi-Instance Learning to enable slide-level prediction given only slide-level labels. Among these models, vanilla attention mechanisms without pairwise interactions have traditionally been employed but are unable to model contextual information. More recently, self-attention models have been utilized to address this issue. To alleviate the computational complexity of long sequences in large WSIs, methods like HIPT use region-slicing, and TransMIL employs approximation of full self-attention. Both approaches suffer from suboptimal performance due to the loss of key information. Moreover, their use of absolute positional embedding struggles to effectively handle long contextual dependencies in shape-varying WSIs. In this paper, we first analyze how the low-rank nature of the long-sequence attention matrix constrains the representation ability of WSI modelling. Then, we demonstrate that the rank of attention matrix can be improved by focusing on local interactions via a local attention mask. Our analysis shows that the local mask aligns with the attention patterns in the lower layers of the Transformer. Furthermore, the local attention mask can be implemented during chunked attention calculation, reducing the quadratic computational complexity to linear with a small local bandwidth. Building on this, we propose a local-global hybrid Transformer for both computational acceleration and local-global information interactions modelling. Our method, Long-contextual MIL (LongMIL), is evaluated through extensive experiments on various WSI tasks to validate its superiority. Our code will be available at github.com/invoker-LL/Long-MIL.
Large-scale cervical precancerous screening via AI-assisted cytology whole slide image analysis
Jul 28, 2024



Abstract:Cervical Cancer continues to be the leading gynecological malignancy, posing a persistent threat to women's health on a global scale. Early screening via cytology Whole Slide Image (WSI) diagnosis is critical to prevent this Cancer progression and improve survival rate, but pathologist's single test suffers inevitable false negative due to the immense number of cells that need to be reviewed within a WSI. Though computer-aided automated diagnostic models can serve as strong complement for pathologists, their effectiveness is hampered by the paucity of extensive and detailed annotations, coupled with the limited interpretability and robustness. These factors significantly hinder their practical applicability and reliability in clinical settings. To tackle these challenges, we develop an AI approach, which is a Scalable Technology for Robust and Interpretable Diagnosis built on Extensive data (STRIDE) of cervical cytology. STRIDE addresses the bottleneck of limited annotations by integrating patient-level labels with a small portion of cell-level labels through an end-to-end training strategy, facilitating scalable learning across extensive datasets. To further improve the robustness to real-world domain shifts of cytology slide-making and imaging, STRIDE employs color adversarial samples training that mimic staining and imaging variations. Lastly, to achieve pathologist-level interpretability for the trustworthiness in clinical settings, STRIDE can generate explanatory textual descriptions that simulates pathologists' diagnostic processes by cell image feature and textual description alignment. Conducting extensive experiments and evaluations in 183 medical centers with a dataset of 341,889 WSIs and 0.1 billion cells from cervical cytology patients, STRIDE has demonstrated a remarkable superiority over previous state-of-the-art techniques.
WSI-VQA: Interpreting Whole Slide Images by Generative Visual Question Answering
Jul 08, 2024



Abstract:Whole slide imaging is routinely adopted for carcinoma diagnosis and prognosis. Abundant experience is required for pathologists to achieve accurate and reliable diagnostic results of whole slide images (WSI). The huge size and heterogeneous features of WSIs make the workflow of pathological reading extremely time-consuming. In this paper, we propose a novel framework (WSI-VQA) to interpret WSIs by generative visual question answering. WSI-VQA shows universality by reframing various kinds of slide-level tasks in a question-answering pattern, in which pathologists can achieve immunohistochemical grading, survival prediction, and tumor subtyping following human-machine interaction. Furthermore, we establish a WSI-VQA dataset which contains 8672 slide-level question-answering pairs with 977 WSIs. Besides the ability to deal with different slide-level tasks, our generative model which is named Wsi2Text Transformer (W2T) outperforms existing discriminative models in medical correctness, which reveals the potential of our model to be applied in the clinical scenario. Additionally, we also visualize the co-attention mapping between word embeddings and WSIs as an intuitive explanation for diagnostic results. The dataset and related code are available at https://github.com/cpystan/WSI-VQA.
TikTokActions: A TikTok-Derived Video Dataset for Human Action Recognition
Feb 14, 2024



Abstract:The increasing variety and quantity of tagged multimedia content on platforms such as TikTok provides an opportunity to advance computer vision modeling. We have curated a distinctive dataset of 283,582 unique video clips categorized under 386 hashtags relating to modern human actions. We release this dataset as a valuable resource for building domain-specific foundation models for human movement modeling tasks such as action recognition. To validate this dataset, which we name TikTokActions, we perform two sets of experiments. First, we pretrain the state-of-the-art VideoMAEv2 with a ViT-base backbone on TikTokActions subset, and then fine-tune and evaluate on popular datasets such as UCF101 and the HMDB51. We find that the performance of the model pre-trained using our Tik-Tok dataset is comparable to models trained on larger action recognition datasets (95.3% on UCF101 and 53.24% on HMDB51). Furthermore, our investigation into the relationship between pre-training dataset size and fine-tuning performance reveals that beyond a certain threshold, the incremental benefit of larger training sets diminishes. This work introduces a useful TikTok video dataset that is available for public use and provides insights into the marginal benefit of increasing pre-training dataset sizes for video-based foundation models.
Benchmarking PathCLIP for Pathology Image Analysis
Jan 05, 2024
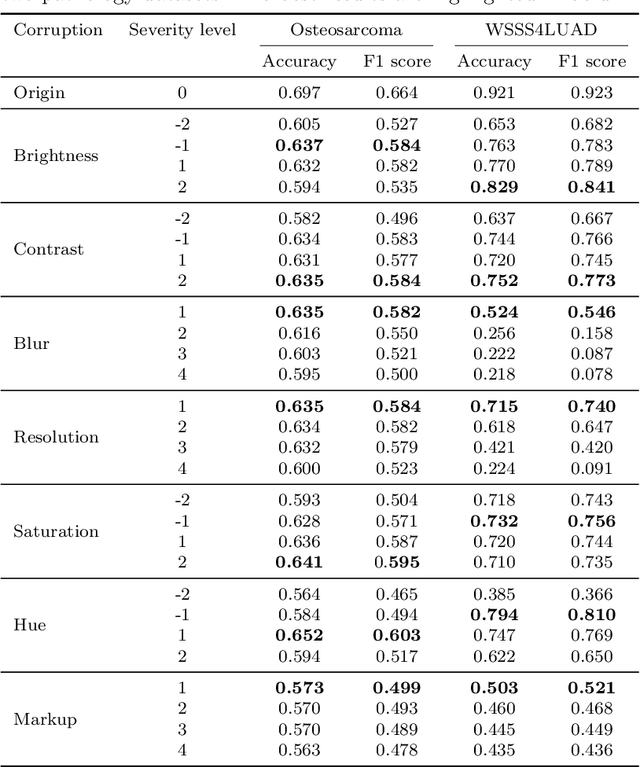

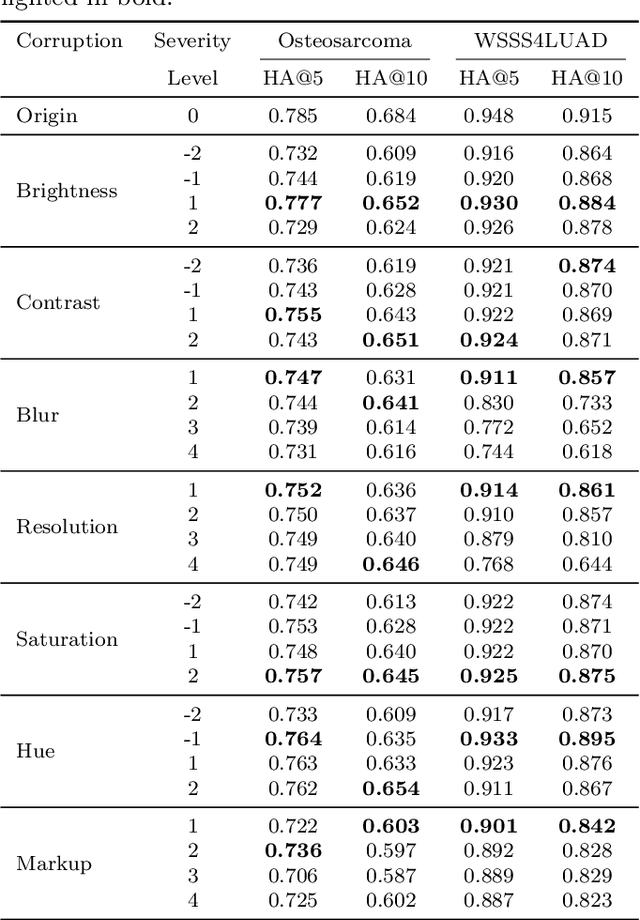
Abstract:Accurate image classification and retrieval are of importance for clinical diagnosis and treatment decision-making. The recent contrastive language-image pretraining (CLIP) model has shown remarkable proficiency in understanding natural images. Drawing inspiration from CLIP, PathCLIP is specifically designed for pathology image analysis, utilizing over 200,000 image and text pairs in training. While the performance the PathCLIP is impressive, its robustness under a wide range of image corruptions remains unknown. Therefore, we conduct an extensive evaluation to analyze the performance of PathCLIP on various corrupted images from the datasets of Osteosarcoma and WSSS4LUAD. In our experiments, we introduce seven corruption types including brightness, contrast, Gaussian blur, resolution, saturation, hue, and markup at four severity levels. Through experiments, we find that PathCLIP is relatively robustness to image corruptions and surpasses OpenAI-CLIP and PLIP in zero-shot classification. Among the seven corruptions, blur and resolution can cause server performance degradation of the PathCLIP. This indicates that ensuring the quality of images is crucial before conducting a clinical test. Additionally, we assess the robustness of PathCLIP in the task of image-image retrieval, revealing that PathCLIP performs less effectively than PLIP on Osteosarcoma but performs better on WSSS4LUAD under diverse corruptions. Overall, PathCLIP presents impressive zero-shot classification and retrieval performance for pathology images, but appropriate care needs to be taken when using it. We hope this study provides a qualitative impression of PathCLIP and helps understand its differences from other CLIP models.
MI-Gen: Multiple Instance Generation of Pathology Reports for Gigapixel Whole-Slide Images
Nov 27, 2023



Abstract:Whole slide images are the foundation of digital pathology for the diagnosis and treatment of carcinomas. Writing pathology reports is laborious and error-prone for inexperienced pathologists. To reduce the workload and improve clinical automation, we investigate how to generate pathology reports given whole slide images. On the data end, we curated the largest WSI-text dataset (TCGA-PathoText). In specific, we collected nearly 10000 high-quality WSI-text pairs for visual-language models by recognizing and cleaning pathology reports which narrate diagnostic slides in TCGA. On the model end, we propose the multiple instance generative model (MI-Gen) which can produce pathology reports for gigapixel WSIs. We benchmark our model on the largest subset of TCGA-PathoText. Experimental results show our model can generate pathology reports which contain multiple clinical clues. Furthermore, WSI-text prediction can be seen as an approach of visual-language pre-training, which enables our model to be transferred to downstream diagnostic tasks like carcinoma grading and phenotyping. We observe that simple semantic extraction from the pathology reports can achieve the best performance (0.838 of F1 score) on BRCA subtyping without adding extra parameters or tricky fine-tuning. Our collected dataset and related code will all be publicly available.
Exploring Unsupervised Cell Recognition with Prior Self-activation Maps
Aug 22, 2023



Abstract:The success of supervised deep learning models on cell recognition tasks relies on detailed annotations. Many previous works have managed to reduce the dependency on labels. However, considering the large number of cells contained in a patch, costly and inefficient labeling is still inevitable. To this end, we explored label-free methods for cell recognition. Prior self-activation maps (PSM) are proposed to generate pseudo masks as training targets. To be specific, an activation network is trained with self-supervised learning. The gradient information in the shallow layers of the network is aggregated to generate prior self-activation maps. Afterward, a semantic clustering module is then introduced as a pipeline to transform PSMs to pixel-level semantic pseudo masks for downstream tasks. We evaluated our method on two histological datasets: MoNuSeg (cell segmentation) and BCData (multi-class cell detection). Compared with other fully-supervised and weakly-supervised methods, our method can achieve competitive performance without any manual annotations. Our simple but effective framework can also achieve multi-class cell detection which can not be done by existing unsupervised methods. The results show the potential of PSMs that might inspire other research to deal with the hunger for labels in medical area.
Unsupervised Dense Nuclei Detection and Segmentation with Prior Self-activation Map For Histology Images
Oct 14, 2022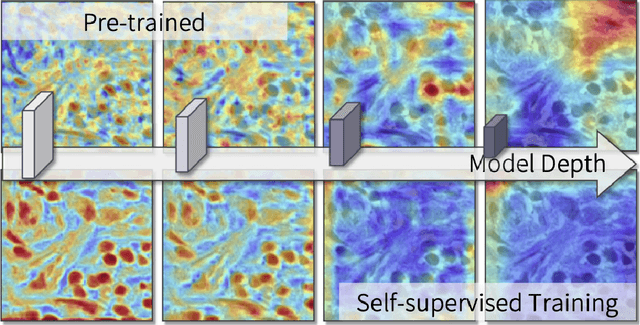
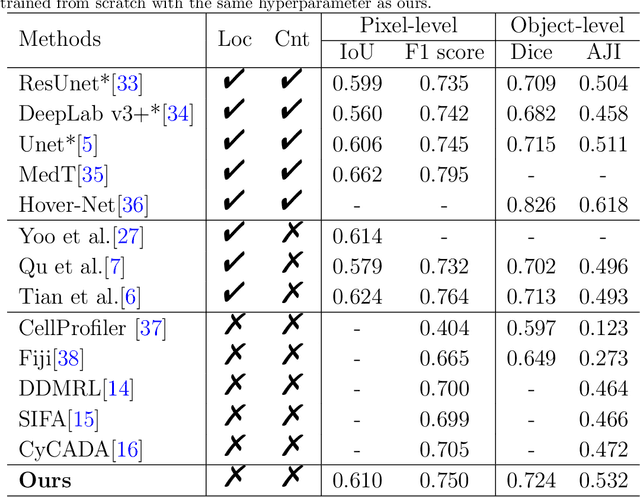
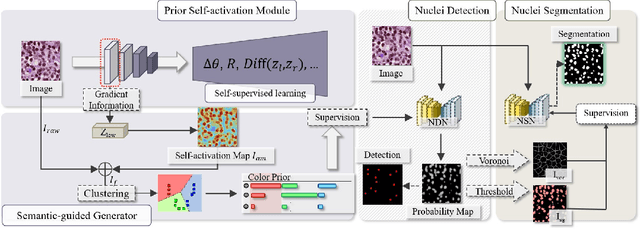
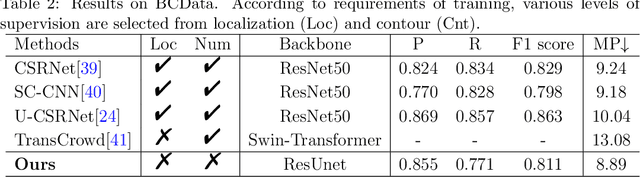
Abstract:The success of supervised deep learning models in medical image segmentation relies on detailed annotations. However, labor-intensive manual labeling is costly and inefficient, especially in dense object segmentation. To this end, we propose a self-supervised learning based approach with a Prior Self-activation Module (PSM) that generates self-activation maps from the input images to avoid labeling costs and further produce pseudo masks for the downstream task. To be specific, we firstly train a neural network using self-supervised learning and utilize the gradient information in the shallow layers of the network to generate self-activation maps. Afterwards, a semantic-guided generator is then introduced as a pipeline to transform visual representations from PSM to pixel-level semantic pseudo masks for downstream tasks. Furthermore, a two-stage training module, consisting of a nuclei detection network and a nuclei segmentation network, is adopted to achieve the final segmentation. Experimental results show the effectiveness on two public pathological datasets. Compared with other fully-supervised and weakly-supervised methods, our method can achieve competitive performance without any manual annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge