Zhaoxiang Ye
Benchmarking PathCLIP for Pathology Image Analysis
Jan 05, 2024
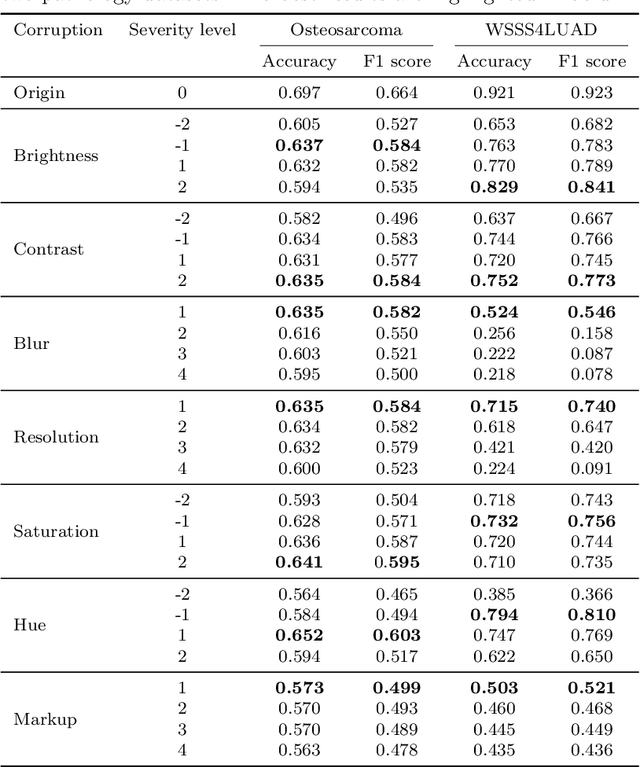

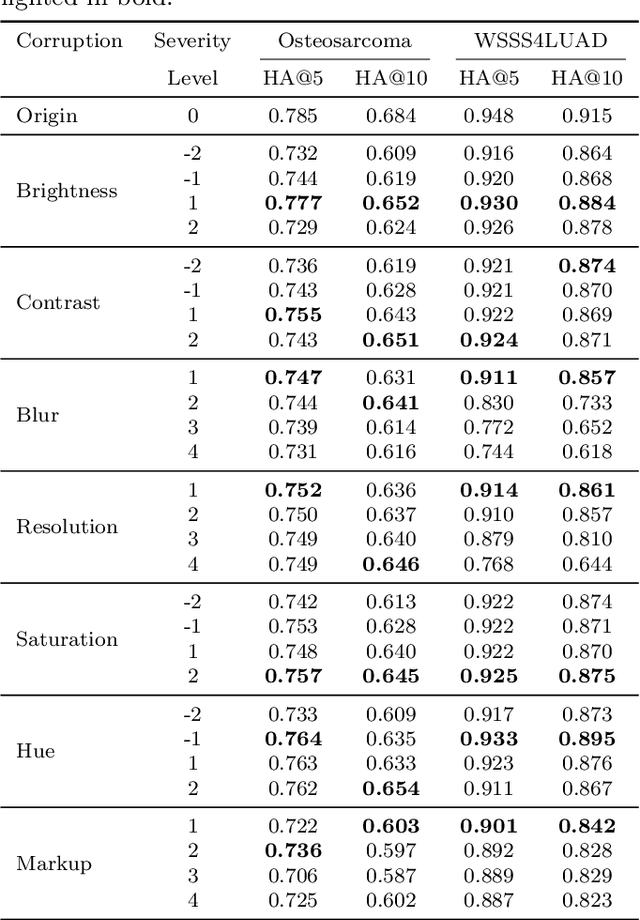
Abstract:Accurate image classification and retrieval are of importance for clinical diagnosis and treatment decision-making. The recent contrastive language-image pretraining (CLIP) model has shown remarkable proficiency in understanding natural images. Drawing inspiration from CLIP, PathCLIP is specifically designed for pathology image analysis, utilizing over 200,000 image and text pairs in training. While the performance the PathCLIP is impressive, its robustness under a wide range of image corruptions remains unknown. Therefore, we conduct an extensive evaluation to analyze the performance of PathCLIP on various corrupted images from the datasets of Osteosarcoma and WSSS4LUAD. In our experiments, we introduce seven corruption types including brightness, contrast, Gaussian blur, resolution, saturation, hue, and markup at four severity levels. Through experiments, we find that PathCLIP is relatively robustness to image corruptions and surpasses OpenAI-CLIP and PLIP in zero-shot classification. Among the seven corruptions, blur and resolution can cause server performance degradation of the PathCLIP. This indicates that ensuring the quality of images is crucial before conducting a clinical test. Additionally, we assess the robustness of PathCLIP in the task of image-image retrieval, revealing that PathCLIP performs less effectively than PLIP on Osteosarcoma but performs better on WSSS4LUAD under diverse corruptions. Overall, PathCLIP presents impressive zero-shot classification and retrieval performance for pathology images, but appropriate care needs to be taken when using it. We hope this study provides a qualitative impression of PathCLIP and helps understand its differences from other CLIP models.
Low-Dose CT Denoising Using a Structure-Preserving Kernel Prediction Network
May 31, 2021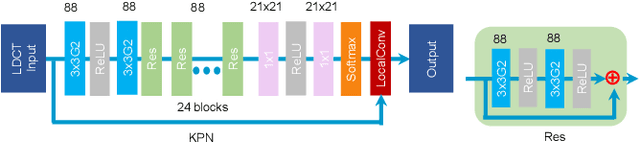
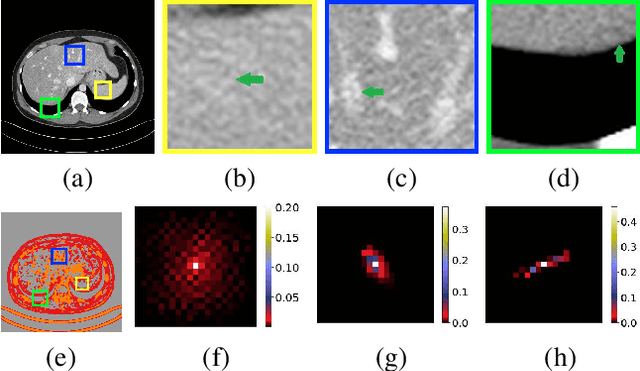
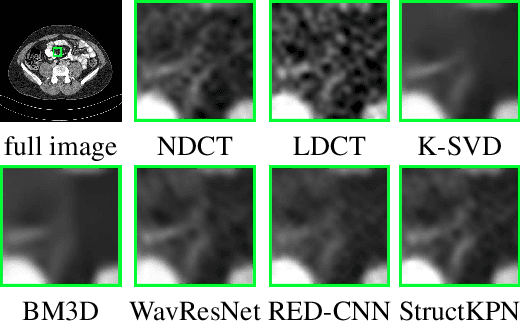

Abstract:Low-dose CT has been a key diagnostic imaging modality to reduce the potential risk of radiation overdose to patient health. Despite recent advances, CNN-based approaches typically apply filters in a spatially invariant way and adopt similar pixel-level losses, which treat all regions of the CT image equally and can be inefficient when fine-grained structures coexist with non-uniformly distributed noises. To address this issue, we propose a Structure-preserving Kernel Prediction Network (StructKPN) that combines the kernel prediction network with a structure-aware loss function that utilizes the pixel gradient statistics and guides the model towards spatially-variant filters that enhance noise removal, prevent over-smoothing and preserve detailed structures for different regions in CT imaging. Extensive experiments demonstrated that our approach achieved superior performance on both synthetic and non-synthetic datasets, and better preserves structures that are highly desired in clinical screening and low-dose protocol optimization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge