Xueping Jing
Benchmarking PathCLIP for Pathology Image Analysis
Jan 05, 2024
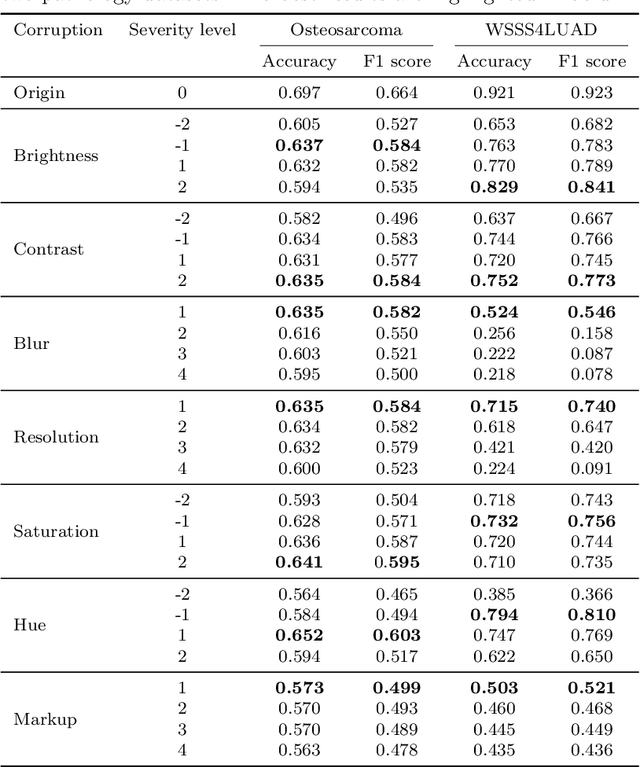

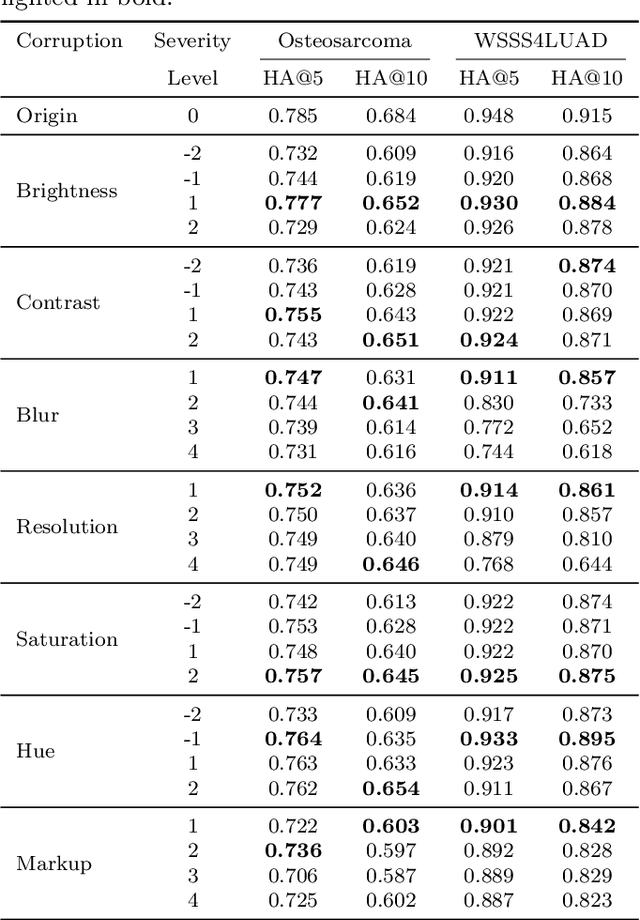
Abstract:Accurate image classification and retrieval are of importance for clinical diagnosis and treatment decision-making. The recent contrastive language-image pretraining (CLIP) model has shown remarkable proficiency in understanding natural images. Drawing inspiration from CLIP, PathCLIP is specifically designed for pathology image analysis, utilizing over 200,000 image and text pairs in training. While the performance the PathCLIP is impressive, its robustness under a wide range of image corruptions remains unknown. Therefore, we conduct an extensive evaluation to analyze the performance of PathCLIP on various corrupted images from the datasets of Osteosarcoma and WSSS4LUAD. In our experiments, we introduce seven corruption types including brightness, contrast, Gaussian blur, resolution, saturation, hue, and markup at four severity levels. Through experiments, we find that PathCLIP is relatively robustness to image corruptions and surpasses OpenAI-CLIP and PLIP in zero-shot classification. Among the seven corruptions, blur and resolution can cause server performance degradation of the PathCLIP. This indicates that ensuring the quality of images is crucial before conducting a clinical test. Additionally, we assess the robustness of PathCLIP in the task of image-image retrieval, revealing that PathCLIP performs less effectively than PLIP on Osteosarcoma but performs better on WSSS4LUAD under diverse corruptions. Overall, PathCLIP presents impressive zero-shot classification and retrieval performance for pathology images, but appropriate care needs to be taken when using it. We hope this study provides a qualitative impression of PathCLIP and helps understand its differences from other CLIP models.
Efficient convolutional neural networks for multi-planar lung nodule detection: improvement on small nodule identification
Jan 13, 2020



Abstract:We propose a multi-planar pulmonary nodule detection system using convolutional neural networks. The 2-D convolutional neural network model, U-net++, was trained by axial, coronal, and sagittal slices for the candidate detection task. All possible nodule candidates from the three different planes are combined. For false positive reduction, we apply 3-D multi-scale dense convolutional neural networks to efficiently remove false positive candidates. We use the public LIDC-IDRI dataset which includes 888 CT scans with 1186 nodules annotated by four radiologists. After ten-fold cross-validation, our proposed system achieves a sensitivity of 95.3% with 0.5 false positive/scan and a sensitivity of 96.2% with 1.0 false positive/scan. Although it is difficult to detect small nodules (i.e. nodules with a diameter < 6 mm), our designed CAD system reaches a sensitivity of 93.8% (94.6%) of these small nodules at an overall false positive rate of 0.5 (1.0) false positives/scan. At the nodule candidate detection stage, the proposed system detected 98.1% of nodules after merging the predictions from all three planes. Using only the 1 mm axial slices resulted in the detection of 91.1% of nodules, which is better than that of utilizing solely the coronal or sagittal slices. The results show that a multi-planar method is capable to detect more nodules compared to using a single plane. Our approach achieves state-of-the-art performance on this dataset, which demonstrates the effectiveness and efficiency of our developed CAD system for lung nodule detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge