Lin Yang
Tony
CompactRAG: Reducing LLM Calls and Token Overhead in Multi-Hop Question Answering
Feb 05, 2026Abstract:Retrieval-augmented generation (RAG) has become a key paradigm for knowledge-intensive question answering. However, existing multi-hop RAG systems remain inefficient, as they alternate between retrieval and reasoning at each step, resulting in repeated LLM calls, high token consumption, and unstable entity grounding across hops. We propose CompactRAG, a simple yet effective framework that decouples offline corpus restructuring from online reasoning. In the offline stage, an LLM reads the corpus once and converts it into an atomic QA knowledge base, which represents knowledge as minimal, fine-grained question-answer pairs. In the online stage, complex queries are decomposed and carefully rewritten to preserve entity consistency, and are resolved through dense retrieval followed by RoBERTa-based answer extraction. Notably, during inference, the LLM is invoked only twice in total - once for sub-question decomposition and once for final answer synthesis - regardless of the number of reasoning hops. Experiments on HotpotQA, 2WikiMultiHopQA, and MuSiQue demonstrate that CompactRAG achieves competitive accuracy while substantially reducing token consumption compared to iterative RAG baselines, highlighting a cost-efficient and practical approach to multi-hop reasoning over large knowledge corpora. The implementation is available at GitHub.
Factored Causal Representation Learning for Robust Reward Modeling in RLHF
Jan 29, 2026Abstract:A reliable reward model is essential for aligning large language models with human preferences through reinforcement learning from human feedback. However, standard reward models are susceptible to spurious features that are not causally related to human labels. This can lead to reward hacking, where high predicted reward does not translate into better behavior. In this work, we address this problem from a causal perspective by proposing a factored representation learning framework that decomposes the model's contextual embedding into (1) causal factors that are sufficient for reward prediction and (2) non-causal factors that capture reward-irrelevant attributes such as length or sycophantic bias. The reward head is then constrained to depend only on the causal component. In addition, we introduce an adversarial head trained to predict reward from the non-causal factors, while applying gradient reversal to discourage them from encoding reward-relevant information. Experiments on both mathematical and dialogue tasks demonstrate that our method learns more robust reward models and consistently improves downstream RLHF performance over state-of-the-art baselines. Analyses on length and sycophantic bias further validate the effectiveness of our method in mitigating reward hacking behaviors.
OpenAI GPT-5 System Card
Dec 19, 2025Abstract:This is the system card published alongside the OpenAI GPT-5 launch, August 2025. GPT-5 is a unified system with a smart and fast model that answers most questions, a deeper reasoning model for harder problems, and a real-time router that quickly decides which model to use based on conversation type, complexity, tool needs, and explicit intent (for example, if you say 'think hard about this' in the prompt). The router is continuously trained on real signals, including when users switch models, preference rates for responses, and measured correctness, improving over time. Once usage limits are reached, a mini version of each model handles remaining queries. This system card focuses primarily on gpt-5-thinking and gpt-5-main, while evaluations for other models are available in the appendix. The GPT-5 system not only outperforms previous models on benchmarks and answers questions more quickly, but -- more importantly -- is more useful for real-world queries. We've made significant advances in reducing hallucinations, improving instruction following, and minimizing sycophancy, and have leveled up GPT-5's performance in three of ChatGPT's most common uses: writing, coding, and health. All of the GPT-5 models additionally feature safe-completions, our latest approach to safety training to prevent disallowed content. Similarly to ChatGPT agent, we have decided to treat gpt-5-thinking as High capability in the Biological and Chemical domain under our Preparedness Framework, activating the associated safeguards. While we do not have definitive evidence that this model could meaningfully help a novice to create severe biological harm -- our defined threshold for High capability -- we have chosen to take a precautionary approach.
CodeDance: A Dynamic Tool-integrated MLLM for Executable Visual Reasoning
Dec 19, 2025Abstract:Recent releases such as o3 highlight human-like "thinking with images" reasoning that combines structured tool use with stepwise verification, yet most open-source approaches still rely on text-only chains, rigid visual schemas, or single-step pipelines, limiting flexibility, interpretability, and transferability on complex tasks. We introduce CodeDance, which explores executable code as a general solver for visual reasoning. Unlike fixed-schema calls (e.g., only predicting bounding-box coordinates), CodeDance defines, composes, and executes code to orchestrate multiple tools, compute intermediate results, and render visual artifacts (e.g., boxes, lines, plots) that support transparent, self-checkable reasoning. To guide this process, we introduce a reward for balanced and adaptive tool-call, which balances exploration with efficiency and mitigates tool overuse. Interestingly, beyond the expected capabilities taught by atomic supervision, we empirically observe novel emergent behaviors during RL training: CodeDance demonstrates novel tool invocations, unseen compositions, and cross-task transfer. These behaviors arise without task-specific fine-tuning, suggesting a general and scalable mechanism of executable visual reasoning. Extensive experiments across reasoning benchmarks (e.g., visual search, math, chart QA) show that CodeDance not only consistently outperforms schema-driven and text-only baselines, but also surpasses advanced closed models such as GPT-4o and larger open-source models.
RecGPT-V2 Technical Report
Dec 16, 2025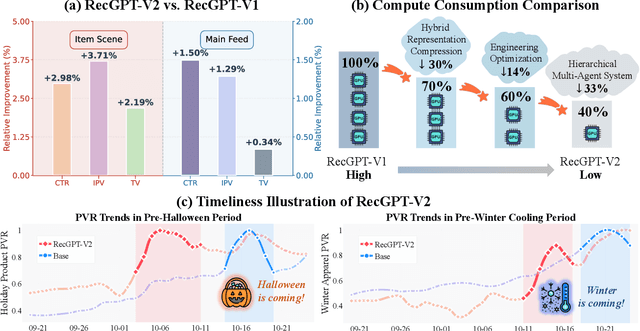
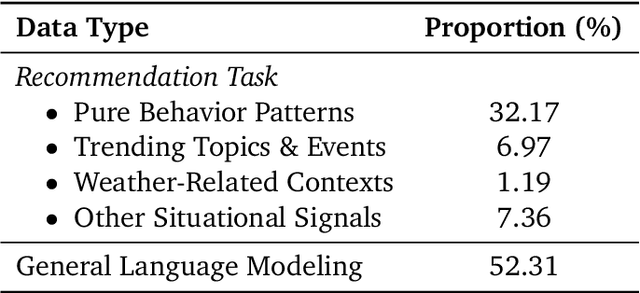
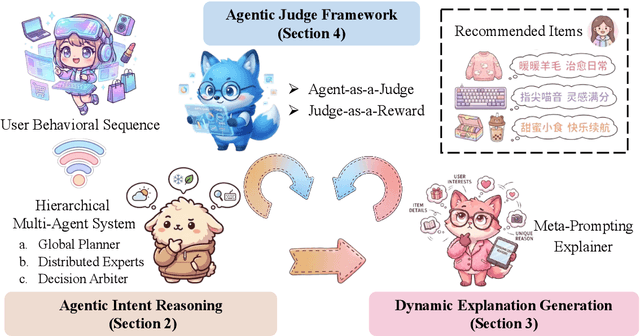
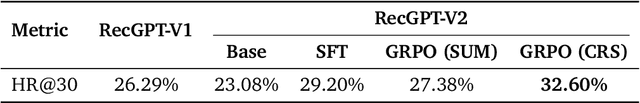
Abstract:Large language models (LLMs) have demonstrated remarkable potential in transforming recommender systems from implicit behavioral pattern matching to explicit intent reasoning. While RecGPT-V1 successfully pioneered this paradigm by integrating LLM-based reasoning into user interest mining and item tag prediction, it suffers from four fundamental limitations: (1) computational inefficiency and cognitive redundancy across multiple reasoning routes; (2) insufficient explanation diversity in fixed-template generation; (3) limited generalization under supervised learning paradigms; and (4) simplistic outcome-focused evaluation that fails to match human standards. To address these challenges, we present RecGPT-V2 with four key innovations. First, a Hierarchical Multi-Agent System restructures intent reasoning through coordinated collaboration, eliminating cognitive duplication while enabling diverse intent coverage. Combined with Hybrid Representation Inference that compresses user-behavior contexts, our framework reduces GPU consumption by 60% and improves exclusive recall from 9.39% to 10.99%. Second, a Meta-Prompting framework dynamically generates contextually adaptive prompts, improving explanation diversity by +7.3%. Third, constrained reinforcement learning mitigates multi-reward conflicts, achieving +24.1% improvement in tag prediction and +13.0% in explanation acceptance. Fourth, an Agent-as-a-Judge framework decomposes assessment into multi-step reasoning, improving human preference alignment. Online A/B tests on Taobao demonstrate significant improvements: +2.98% CTR, +3.71% IPV, +2.19% TV, and +11.46% NER. RecGPT-V2 establishes both the technical feasibility and commercial viability of deploying LLM-powered intent reasoning at scale, bridging the gap between cognitive exploration and industrial utility.
A Specialized Large Language Model for Clinical Reasoning and Diagnosis in Rare Diseases
Nov 18, 2025Abstract:Rare diseases affect hundreds of millions worldwide, yet diagnosis often spans years. Convectional pipelines decouple noisy evidence extraction from downstream inferential diagnosis, and general/medical large language models (LLMs) face scarce real world electronic health records (EHRs), stale domain knowledge, and hallucinations. We assemble a large, domain specialized clinical corpus and a clinician validated reasoning set, and develop RareSeek R1 via staged instruction tuning, chain of thought learning, and graph grounded retrieval. Across multicenter EHR narratives and public benchmarks, RareSeek R1 attains state of the art accuracy, robust generalization, and stability under noisy or overlapping phenotypes. Augmented retrieval yields the largest gains when narratives pair with prioritized variants by resolving ambiguity and aligning candidates to mechanisms. Human studies show performance on par with experienced physicians and consistent gains in assistive use. Notably, transparent reasoning highlights decisive non phenotypic evidence (median 23.1%, such as imaging, interventions, functional tests) underpinning many correct diagnoses. This work advances a narrative first, knowledge integrated reasoning paradigm that shortens the diagnostic odyssey and enables auditable, clinically translatable decision support.
Distributed Multi-Agent Bandits Over Erdős-Rényi Random Networks
Oct 26, 2025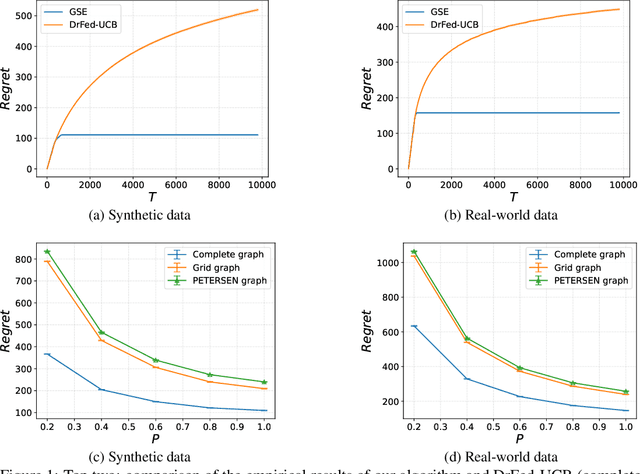
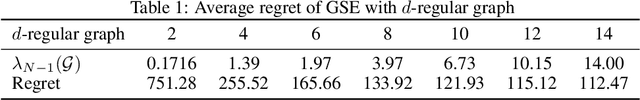
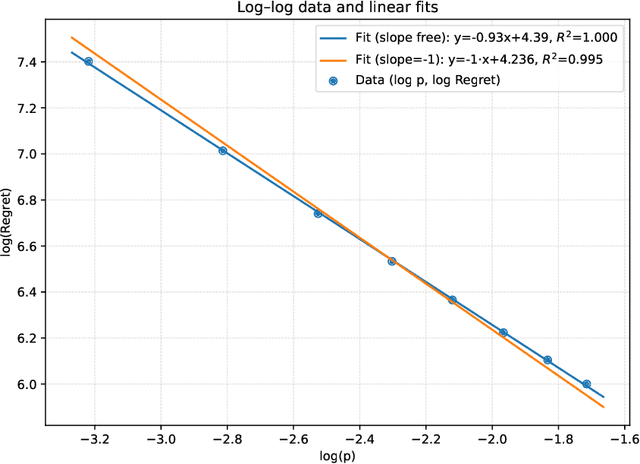
Abstract:We study the distributed multi-agent multi-armed bandit problem with heterogeneous rewards over random communication graphs. Uniquely, at each time step $t$ agents communicate over a time-varying random graph $G_t$ generated by applying the Erd\H{o}s-R\'enyi model to a fixed connected base graph $G$ (for classical Erd\H{o}s-R\'enyi graphs, $G$ is a complete graph), where each potential edge in $G$ is randomly and independently present with the link probability $p$. Notably, the resulting random graph is not necessarily connected at each time step. Each agent's arm rewards follow time-invariant distributions, and the reward distribution for the same arm may differ across agents. The goal is to minimize the cumulative expected regret relative to the global mean reward of each arm, defined as the average of that arm's mean rewards across all agents. To this end, we propose a fully distributed algorithm that integrates the arm elimination strategy with the random gossip algorithm. We theoretically show that the regret upper bound is of order $\log T$ and is highly interpretable, where $T$ is the time horizon. It includes the optimal centralized regret $O\left(\sum_{k: \Delta_k>0} \frac{\log T}{\Delta_k}\right)$ and an additional term $O\left(\frac{N^2 \log T}{p \lambda_{N-1}(Lap(G))} + \frac{KN^2 \log T}{p}\right)$ where $N$ and $K$ denote the total number of agents and arms, respectively. This term reflects the impact of $G$'s algebraic connectivity $\lambda_{N-1}(Lap(G))$ and the link probability $p$, and thus highlights a fundamental trade-off between communication efficiency and regret. As a by-product, we show a nearly optimal regret lower bound. Finally, our numerical experiments not only show the superiority of our algorithm over existing benchmarks, but also validate the theoretical regret scaling with problem complexity.
Clarifying Semantics of In-Context Examples for Unit Test Generation
Oct 02, 2025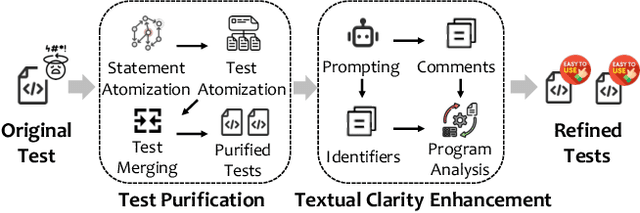
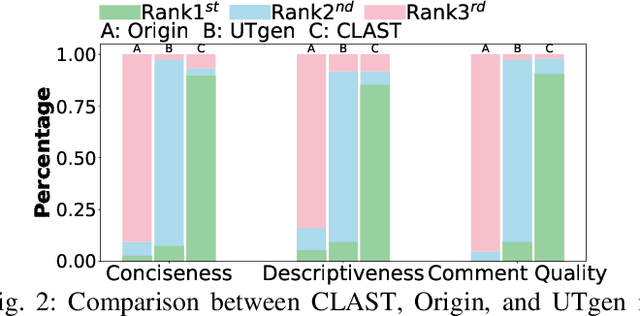
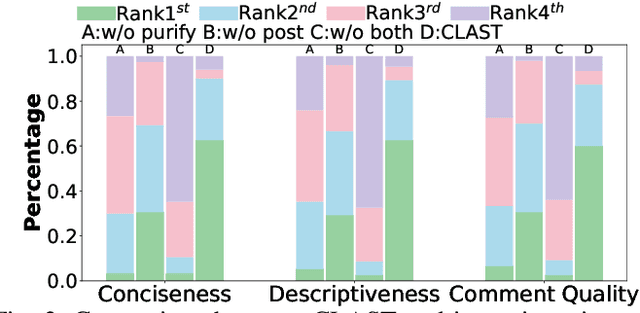
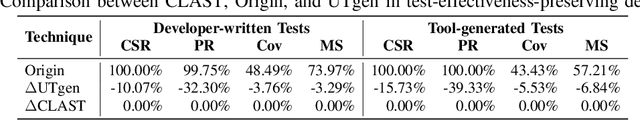
Abstract:Recent advances in large language models (LLMs) have enabled promising performance in unit test generation through in-context learning (ICL). However, the quality of in-context examples significantly influences the effectiveness of generated tests-poorly structured or semantically unclear test examples often lead to suboptimal outputs. In this paper, we propose CLAST, a novel technique that systematically refines unit tests to improve their semantic clarity, thereby enhancing their utility as in-context examples. The approach decomposes complex tests into logically clearer ones and improves semantic clarity through a combination of program analysis and LLM-based rewriting. We evaluated CLAST on four open-source and three industrial projects. The results demonstrate that CLAST largely outperforms UTgen, the state-of-the-art refinement technique, in both preserving test effectiveness and enhancing semantic clarity. Specifically, CLAST fully retains the original effectiveness of unit tests, while UTgen reduces compilation success rate (CSR), pass rate (PR), test coverage (Cov), and mutation score (MS) by an average of 12.90%, 35.82%, 4.65%, and 5.07%, respectively. Over 85.33% of participants in our user study preferred the semantic clarity of CLAST-refined tests. Notably, incorporating CLAST-refined tests as examples effectively improves ICL-based unit test generation approaches such as RAGGen and TELPA, resulting in an average increase of 25.97% in CSR, 28.22% in PR, and 45.99% in Cov for generated tests, compared to incorporating UTgen-refined tests. The insights from the follow-up user study not only reinforce CLAST's potential impact in software testing practice but also illuminate avenues for future research.
DEPTH: Hallucination-Free Relation Extraction via Dependency-Aware Sentence Simplification and Two-tiered Hierarchical Refinement
Aug 20, 2025



Abstract:Relation extraction enables the construction of structured knowledge for many downstream applications. While large language models (LLMs) have shown great promise in this domain, most existing methods concentrate on relation classification, which predicts the semantic relation type between a related entity pair. However, we observe that LLMs often struggle to reliably determine whether a relation exists, especially in cases involving complex sentence structures or intricate semantics, which leads to spurious predictions. Such hallucinations can introduce noisy edges in knowledge graphs, compromising the integrity of structured knowledge and downstream reliability. To address these challenges, we propose DEPTH, a framework that integrates Dependency-aware sEntence simPlification and Two-tiered Hierarchical refinement into the relation extraction pipeline. Given a sentence and its candidate entity pairs, DEPTH operates in two stages: (1) the Grounding module extracts relations for each pair by leveraging their shortest dependency path, distilling the sentence into a minimal yet coherent relational context that reduces syntactic noise while preserving key semantics; (2) the Refinement module aggregates all local predictions and revises them based on a holistic understanding of the sentence, correcting omissions and inconsistencies. We further introduce a causality-driven reward model that mitigates reward hacking by disentangling spurious correlations, enabling robust fine-tuning via reinforcement learning with human feedback. Experiments on six benchmarks demonstrate that DEPTH reduces the average hallucination rate to 7.0\% while achieving a 17.2\% improvement in average F1 score over state-of-the-art baselines.
CPathAgent: An Agent-based Foundation Model for Interpretable High-Resolution Pathology Image Analysis Mimicking Pathologists' Diagnostic Logic
May 26, 2025Abstract:Recent advances in computational pathology have led to the emergence of numerous foundation models. However, these approaches fail to replicate the diagnostic process of pathologists, as they either simply rely on general-purpose encoders with multi-instance learning for classification or directly apply multimodal models to generate reports from images. A significant limitation is their inability to emulate the diagnostic logic employed by pathologists, who systematically examine slides at low magnification for overview before progressively zooming in on suspicious regions to formulate comprehensive diagnoses. To address this gap, we introduce CPathAgent, an innovative agent-based model that mimics pathologists' reasoning processes by autonomously executing zoom-in/out and navigation operations across pathology images based on observed visual features. To achieve this, we develop a multi-stage training strategy unifying patch-level, region-level, and whole-slide capabilities within a single model, which is essential for mimicking pathologists, who require understanding and reasoning capabilities across all three scales. This approach generates substantially more detailed and interpretable diagnostic reports compared to existing methods, particularly for huge region understanding. Additionally, we construct an expert-validated PathMMU-HR$^{2}$, the first benchmark for huge region analysis, a critical intermediate scale between patches and whole slides, as diagnosticians typically examine several key regions rather than entire slides at once. Extensive experiments demonstrate that CPathAgent consistently outperforms existing approaches across three scales of benchmarks, validating the effectiveness of our agent-based diagnostic approach and highlighting a promising direction for the future development of computational pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge