Shravya Shetty
Application and Validation of Geospatial Foundation Model Data for the Prediction of Health Facility Programmatic Outputs -- A Case Study in Malawi
Oct 29, 2025



Abstract:The reliability of routine health data in low and middle-income countries (LMICs) is often constrained by reporting delays and incomplete coverage, necessitating the exploration of novel data sources and analytics. Geospatial Foundation Models (GeoFMs) offer a promising avenue by synthesizing diverse spatial, temporal, and behavioral data into mathematical embeddings that can be efficiently used for downstream prediction tasks. This study evaluated the predictive performance of three GeoFM embedding sources - Google Population Dynamics Foundation Model (PDFM), Google AlphaEarth (derived from satellite imagery), and mobile phone call detail records (CDR) - for modeling 15 routine health programmatic outputs in Malawi, and compared their utility to traditional geospatial interpolation methods. We used XGBoost models on data from 552 health catchment areas (January 2021-May 2023), assessing performance with R2, and using an 80/20 training and test data split with 5-fold cross-validation used in training. While predictive performance was mixed, the embedding-based approaches improved upon baseline geostatistical methods in 13 of 15 (87%) indicators tested. A Multi-GeoFM model integrating all three embedding sources produced the most robust predictions, achieving average 5-fold cross validated R2 values for indicators like population density (0.63), new HIV cases (0.57), and child vaccinations (0.47) and test set R2 of 0.64, 0.68, and 0.55, respectively. Prediction was poor for prediction targets with low primary data availability, such as TB and malnutrition cases. These results demonstrate that GeoFM embeddings imbue a modest predictive improvement for select health and demographic outcomes in an LMIC context. We conclude that the integration of multiple GeoFM sources is an efficient and valuable tool for supplementing and strengthening constrained routine health information systems.
CoCa-CXR: Contrastive Captioners Learn Strong Temporal Structures for Chest X-Ray Vision-Language Understanding
Feb 27, 2025Abstract:Vision-language models have proven to be of great benefit for medical image analysis since they learn rich semantics from both images and reports. Prior efforts have focused on better alignment of image and text representations to enhance image understanding. However, though explicit reference to a prior image is common in Chest X-Ray (CXR) reports, aligning progression descriptions with the semantics differences in image pairs remains under-explored. In this work, we propose two components to address this issue. (1) A CXR report processing pipeline to extract temporal structure. It processes reports with a large language model (LLM) to separate the description and comparison contexts, and extracts fine-grained annotations from reports. (2) A contrastive captioner model for CXR, namely CoCa-CXR, to learn how to both describe images and their temporal progressions. CoCa-CXR incorporates a novel regional cross-attention module to identify local differences between paired CXR images. Extensive experiments show the superiority of CoCa-CXR on both progression analysis and report generation compared to previous methods. Notably, on MS-CXR-T progression classification, CoCa-CXR obtains 65.0% average testing accuracy on five pulmonary conditions, outperforming the previous state-of-the-art (SOTA) model BioViL-T by 4.8%. It also achieves a RadGraph F1 of 24.2% on MIMIC-CXR, which is comparable to the Med-Gemini foundation model.
Health AI Developer Foundations
Nov 26, 2024



Abstract:Robust medical Machine Learning (ML) models have the potential to revolutionize healthcare by accelerating clinical research, improving workflows and outcomes, and producing novel insights or capabilities. Developing such ML models from scratch is cost prohibitive and requires substantial compute, data, and time (e.g., expert labeling). To address these challenges, we introduce Health AI Developer Foundations (HAI-DEF), a suite of pre-trained, domain-specific foundation models, tools, and recipes to accelerate building ML for health applications. The models cover various modalities and domains, including radiology (X-rays and computed tomography), histopathology, dermatological imaging, and audio. These models provide domain specific embeddings that facilitate AI development with less labeled data, shorter training times, and reduced computational costs compared to traditional approaches. In addition, we utilize a common interface and style across these models, and prioritize usability to enable developers to integrate HAI-DEF efficiently. We present model evaluations across various tasks and conclude with a discussion of their application and evaluation, covering the importance of ensuring efficacy, fairness, and equity. Finally, while HAI-DEF and specifically the foundation models lower the barrier to entry for ML in healthcare, we emphasize the importance of validation with problem- and population-specific data for each desired usage setting. This technical report will be updated over time as more modalities and features are added.
General Geospatial Inference with a Population Dynamics Foundation Model
Nov 13, 2024



Abstract:Supporting the health and well-being of dynamic populations around the world requires governmental agencies, organizations and researchers to understand and reason over complex relationships between human behavior and local contexts in order to identify high-risk groups and strategically allocate limited resources. Traditional approaches to these classes of problems often entail developing manually curated, task-specific features and models to represent human behavior and the natural and built environment, which can be challenging to adapt to new, or even, related tasks. To address this, we introduce a Population Dynamics Foundation Model (PDFM) that aims to capture the relationships between diverse data modalities and is applicable to a broad range of geospatial tasks. We first construct a geo-indexed dataset for postal codes and counties across the United States, capturing rich aggregated information on human behavior from maps, busyness, and aggregated search trends, and environmental factors such as weather and air quality. We then model this data and the complex relationships between locations using a graph neural network, producing embeddings that can be adapted to a wide range of downstream tasks using relatively simple models. We evaluate the effectiveness of our approach by benchmarking it on 27 downstream tasks spanning three distinct domains: health indicators, socioeconomic factors, and environmental measurements. The approach achieves state-of-the-art performance on all 27 geospatial interpolation tasks, and on 25 out of the 27 extrapolation and super-resolution tasks. We combined the PDFM with a state-of-the-art forecasting foundation model, TimesFM, to predict unemployment and poverty, achieving performance that surpasses fully supervised forecasting. The full set of embeddings and sample code are publicly available for researchers.
Community search signatures as foundation features for human-centered geospatial modeling
Oct 30, 2024



Abstract:Aggregated relative search frequencies offer a unique composite signal reflecting people's habits, concerns, interests, intents, and general information needs, which are not found in other readily available datasets. Temporal search trends have been successfully used in time series modeling across a variety of domains such as infectious diseases, unemployment rates, and retail sales. However, most existing applications require curating specialized datasets of individual keywords, queries, or query clusters, and the search data need to be temporally aligned with the outcome variable of interest. We propose a novel approach for generating an aggregated and anonymized representation of search interest as foundation features at the community level for geospatial modeling. We benchmark these features using spatial datasets across multiple domains. In zip codes with a population greater than 3000 that cover over 95% of the contiguous US population, our models for predicting missing values in a 20% set of holdout counties achieve an average $R^2$ score of 0.74 across 21 health variables, and 0.80 across 6 demographic and environmental variables. Our results demonstrate that these search features can be used for spatial predictions without strict temporal alignment, and that the resulting models outperform spatial interpolation and state of the art methods using satellite imagery features.
Plots Unlock Time-Series Understanding in Multimodal Models
Oct 03, 2024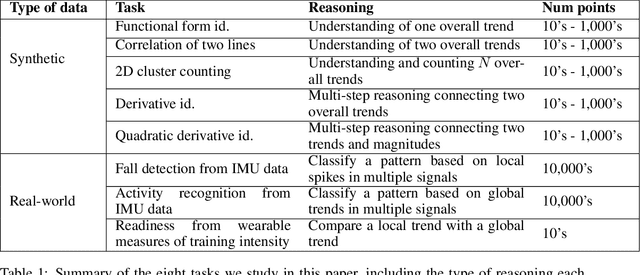

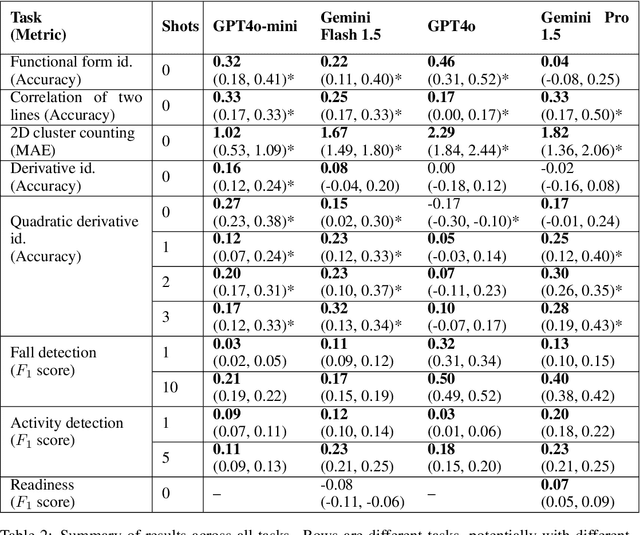
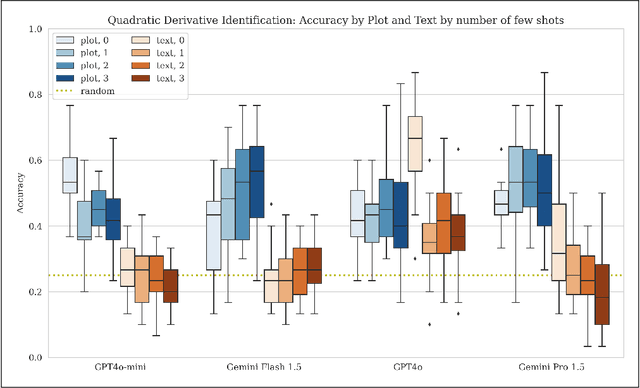
Abstract:While multimodal foundation models can now natively work with data beyond text, they remain underutilized in analyzing the considerable amounts of multi-dimensional time-series data in fields like healthcare, finance, and social sciences, representing a missed opportunity for richer, data-driven insights. This paper proposes a simple but effective method that leverages the existing vision encoders of these models to "see" time-series data via plots, avoiding the need for additional, potentially costly, model training. Our empirical evaluations show that this approach outperforms providing the raw time-series data as text, with the additional benefit that visual time-series representations demonstrate up to a 90% reduction in model API costs. We validate our hypothesis through synthetic data tasks of increasing complexity, progressing from simple functional form identification on clean data, to extracting trends from noisy scatter plots. To demonstrate generalizability from synthetic tasks with clear reasoning steps to more complex, real-world scenarios, we apply our approach to consumer health tasks - specifically fall detection, activity recognition, and readiness assessment - which involve heterogeneous, noisy data and multi-step reasoning. The overall success in plot performance over text performance (up to an 120% performance increase on zero-shot synthetic tasks, and up to 150% performance increase on real-world tasks), across both GPT and Gemini model families, highlights our approach's potential for making the best use of the native capabilities of foundation models.
PathAlign: A vision-language model for whole slide images in histopathology
Jun 27, 2024Abstract:Microscopic interpretation of histopathology images underlies many important diagnostic and treatment decisions. While advances in vision-language modeling raise new opportunities for analysis of such images, the gigapixel-scale size of whole slide images (WSIs) introduces unique challenges. Additionally, pathology reports simultaneously highlight key findings from small regions while also aggregating interpretation across multiple slides, often making it difficult to create robust image-text pairs. As such, pathology reports remain a largely untapped source of supervision in computational pathology, with most efforts relying on region-of-interest annotations or self-supervision at the patch-level. In this work, we develop a vision-language model based on the BLIP-2 framework using WSIs paired with curated text from pathology reports. This enables applications utilizing a shared image-text embedding space, such as text or image retrieval for finding cases of interest, as well as integration of the WSI encoder with a frozen large language model (LLM) for WSI-based generative text capabilities such as report generation or AI-in-the-loop interactions. We utilize a de-identified dataset of over 350,000 WSIs and diagnostic text pairs, spanning a wide range of diagnoses, procedure types, and tissue types. We present pathologist evaluation of text generation and text retrieval using WSI embeddings, as well as results for WSI classification and workflow prioritization (slide-level triaging). Model-generated text for WSIs was rated by pathologists as accurate, without clinically significant error or omission, for 78% of WSIs on average. This work demonstrates exciting potential capabilities for language-aligned WSI embeddings.
Towards a Personal Health Large Language Model
Jun 10, 2024



Abstract:In health, most large language model (LLM) research has focused on clinical tasks. However, mobile and wearable devices, which are rarely integrated into such tasks, provide rich, longitudinal data for personal health monitoring. Here we present Personal Health Large Language Model (PH-LLM), fine-tuned from Gemini for understanding and reasoning over numerical time-series personal health data. We created and curated three datasets that test 1) production of personalized insights and recommendations from sleep patterns, physical activity, and physiological responses, 2) expert domain knowledge, and 3) prediction of self-reported sleep outcomes. For the first task we designed 857 case studies in collaboration with domain experts to assess real-world scenarios in sleep and fitness. Through comprehensive evaluation of domain-specific rubrics, we observed that Gemini Ultra 1.0 and PH-LLM are not statistically different from expert performance in fitness and, while experts remain superior for sleep, fine-tuning PH-LLM provided significant improvements in using relevant domain knowledge and personalizing information for sleep insights. We evaluated PH-LLM domain knowledge using multiple choice sleep medicine and fitness examinations. PH-LLM achieved 79% on sleep and 88% on fitness, exceeding average scores from a sample of human experts. Finally, we trained PH-LLM to predict self-reported sleep quality outcomes from textual and multimodal encoding representations of wearable data, and demonstrate that multimodal encoding is required to match performance of specialized discriminative models. Although further development and evaluation are necessary in the safety-critical personal health domain, these results demonstrate both the broad knowledge and capabilities of Gemini models and the benefit of contextualizing physiological data for personal health applications as done with PH-LLM.
Advancing Multimodal Medical Capabilities of Gemini
May 06, 2024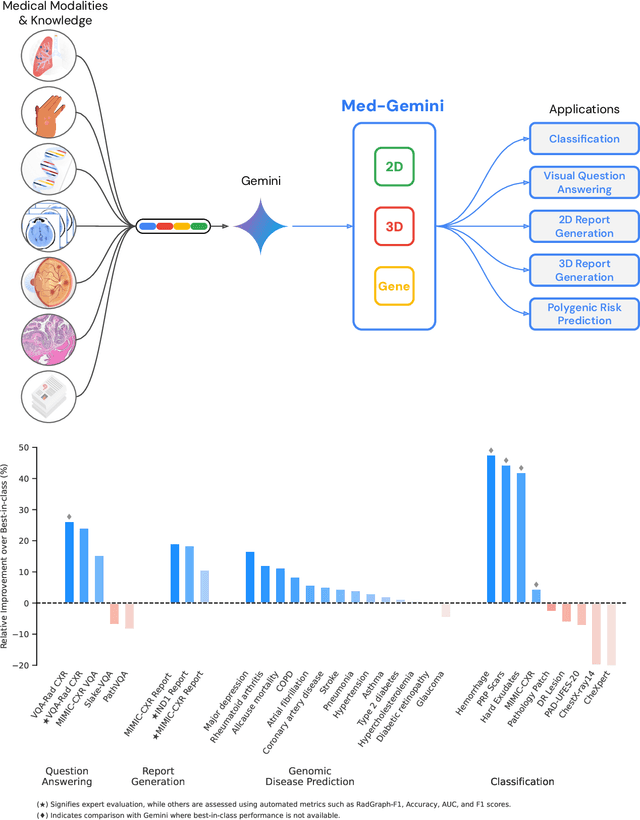
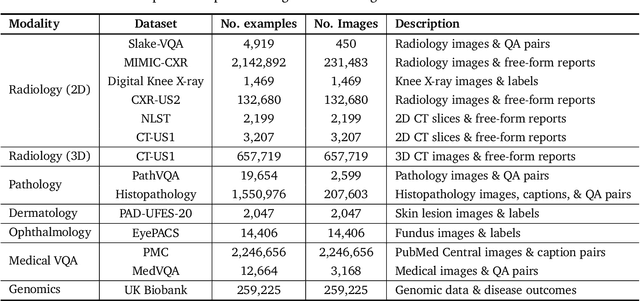

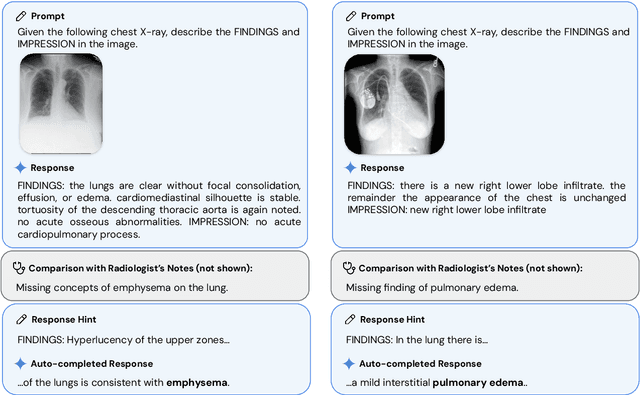
Abstract:Many clinical tasks require an understanding of specialized data, such as medical images and genomics, which is not typically found in general-purpose large multimodal models. Building upon Gemini's multimodal models, we develop several models within the new Med-Gemini family that inherit core capabilities of Gemini and are optimized for medical use via fine-tuning with 2D and 3D radiology, histopathology, ophthalmology, dermatology and genomic data. Med-Gemini-2D sets a new standard for AI-based chest X-ray (CXR) report generation based on expert evaluation, exceeding previous best results across two separate datasets by an absolute margin of 1% and 12%, where 57% and 96% of AI reports on normal cases, and 43% and 65% on abnormal cases, are evaluated as "equivalent or better" than the original radiologists' reports. We demonstrate the first ever large multimodal model-based report generation for 3D computed tomography (CT) volumes using Med-Gemini-3D, with 53% of AI reports considered clinically acceptable, although additional research is needed to meet expert radiologist reporting quality. Beyond report generation, Med-Gemini-2D surpasses the previous best performance in CXR visual question answering (VQA) and performs well in CXR classification and radiology VQA, exceeding SoTA or baselines on 17 of 20 tasks. In histopathology, ophthalmology, and dermatology image classification, Med-Gemini-2D surpasses baselines across 18 out of 20 tasks and approaches task-specific model performance. Beyond imaging, Med-Gemini-Polygenic outperforms the standard linear polygenic risk score-based approach for disease risk prediction and generalizes to genetically correlated diseases for which it has never been trained. Although further development and evaluation are necessary in the safety-critical medical domain, our results highlight the potential of Med-Gemini across a wide range of medical tasks.
HeAR -- Health Acoustic Representations
Mar 04, 2024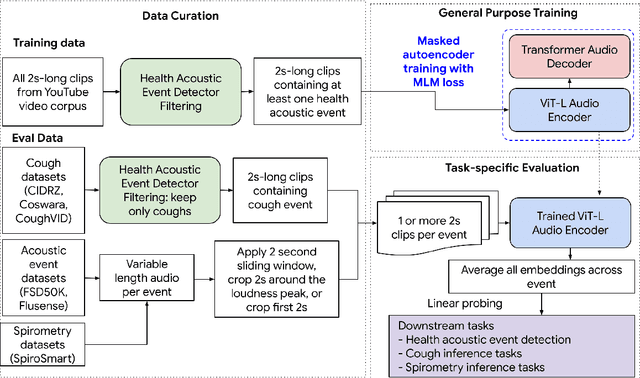
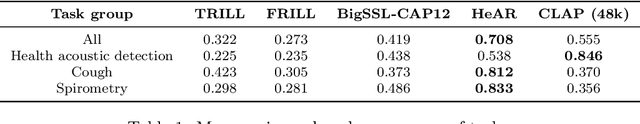
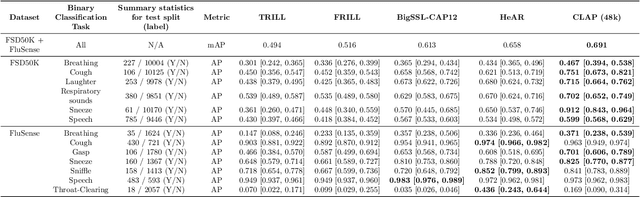
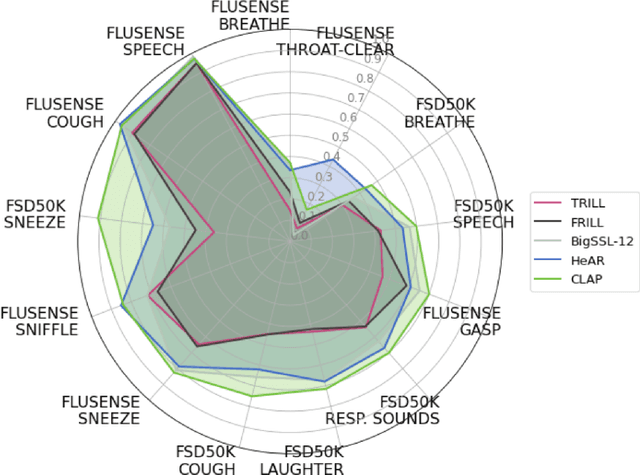
Abstract:Health acoustic sounds such as coughs and breaths are known to contain useful health signals with significant potential for monitoring health and disease, yet are underexplored in the medical machine learning community. The existing deep learning systems for health acoustics are often narrowly trained and evaluated on a single task, which is limited by data and may hinder generalization to other tasks. To mitigate these gaps, we develop HeAR, a scalable self-supervised learning-based deep learning system using masked autoencoders trained on a large dataset of 313 million two-second long audio clips. Through linear probes, we establish HeAR as a state-of-the-art health audio embedding model on a benchmark of 33 health acoustic tasks across 6 datasets. By introducing this work, we hope to enable and accelerate further health acoustics research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge