Marcin Sieniek
General Geospatial Inference with a Population Dynamics Foundation Model
Nov 13, 2024



Abstract:Supporting the health and well-being of dynamic populations around the world requires governmental agencies, organizations and researchers to understand and reason over complex relationships between human behavior and local contexts in order to identify high-risk groups and strategically allocate limited resources. Traditional approaches to these classes of problems often entail developing manually curated, task-specific features and models to represent human behavior and the natural and built environment, which can be challenging to adapt to new, or even, related tasks. To address this, we introduce a Population Dynamics Foundation Model (PDFM) that aims to capture the relationships between diverse data modalities and is applicable to a broad range of geospatial tasks. We first construct a geo-indexed dataset for postal codes and counties across the United States, capturing rich aggregated information on human behavior from maps, busyness, and aggregated search trends, and environmental factors such as weather and air quality. We then model this data and the complex relationships between locations using a graph neural network, producing embeddings that can be adapted to a wide range of downstream tasks using relatively simple models. We evaluate the effectiveness of our approach by benchmarking it on 27 downstream tasks spanning three distinct domains: health indicators, socioeconomic factors, and environmental measurements. The approach achieves state-of-the-art performance on all 27 geospatial interpolation tasks, and on 25 out of the 27 extrapolation and super-resolution tasks. We combined the PDFM with a state-of-the-art forecasting foundation model, TimesFM, to predict unemployment and poverty, achieving performance that surpasses fully supervised forecasting. The full set of embeddings and sample code are publicly available for researchers.
ELIXR: Towards a general purpose X-ray artificial intelligence system through alignment of large language models and radiology vision encoders
Aug 02, 2023Abstract:Our approach, which we call Embeddings for Language/Image-aligned X-Rays, or ELIXR, leverages a language-aligned image encoder combined or grafted onto a fixed LLM, PaLM 2, to perform a broad range of tasks. We train this lightweight adapter architecture using images paired with corresponding free-text radiology reports from the MIMIC-CXR dataset. ELIXR achieved state-of-the-art performance on zero-shot chest X-ray (CXR) classification (mean AUC of 0.850 across 13 findings), data-efficient CXR classification (mean AUCs of 0.893 and 0.898 across five findings (atelectasis, cardiomegaly, consolidation, pleural effusion, and pulmonary edema) for 1% (~2,200 images) and 10% (~22,000 images) training data), and semantic search (0.76 normalized discounted cumulative gain (NDCG) across nineteen queries, including perfect retrieval on twelve of them). Compared to existing data-efficient methods including supervised contrastive learning (SupCon), ELIXR required two orders of magnitude less data to reach similar performance. ELIXR also showed promise on CXR vision-language tasks, demonstrating overall accuracies of 58.7% and 62.5% on visual question answering and report quality assurance tasks, respectively. These results suggest that ELIXR is a robust and versatile approach to CXR AI.
Enabling faster and more reliable sonographic assessment of gestational age through machine learning
Mar 22, 2022



Abstract:Fetal ultrasounds are an essential part of prenatal care and can be used to estimate gestational age (GA). Accurate GA assessment is important for providing appropriate prenatal care throughout pregnancy and identifying complications such as fetal growth disorders. Since derivation of GA from manual fetal biometry measurements (head, abdomen, femur) are operator-dependent and time-consuming, there have been a number of research efforts focused on using artificial intelligence (AI) models to estimate GA using standard biometry images, but there is still room to improve the accuracy and reliability of these AI systems for widescale adoption. To improve GA estimates, without significant change to provider workflows, we leverage AI to interpret standard plane ultrasound images as well as 'fly-to' ultrasound videos, which are 5-10s videos automatically recorded as part of the standard of care before the still image is captured. We developed and validated three AI models: an image model using standard plane images, a video model using fly-to videos, and an ensemble model (combining both image and video). All three were statistically superior to standard fetal biometry-based GA estimates derived by expert sonographers, the ensemble model has the lowest mean absolute error (MAE) compared to the clinical standard fetal biometry (mean difference: -1.51 $\pm$ 3.96 days, 95% CI [-1.9, -1.1]) on a test set that consisted of 404 participants. We showed that our models outperform standard biometry by a more substantial margin on fetuses that were small for GA. Our AI models have the potential to empower trained operators to estimate GA with higher accuracy while reducing the amount of time required and user variability in measurement acquisition.
AI system for fetal ultrasound in low-resource settings
Mar 18, 2022



Abstract:Despite considerable progress in maternal healthcare, maternal and perinatal deaths remain high in low-to-middle income countries. Fetal ultrasound is an important component of antenatal care, but shortage of adequately trained healthcare workers has limited its adoption. We developed and validated an artificial intelligence (AI) system that uses novice-acquired "blind sweep" ultrasound videos to estimate gestational age (GA) and fetal malpresentation. We further addressed obstacles that may be encountered in low-resourced settings. Using a simplified sweep protocol with real-time AI feedback on sweep quality, we have demonstrated the generalization of model performance to minimally trained novice ultrasound operators using low cost ultrasound devices with on-device AI integration. The GA model was non-inferior to standard fetal biometry estimates with as few as two sweeps, and the fetal malpresentation model had high AUC-ROCs across operators and devices. Our AI models have the potential to assist in upleveling the capabilities of lightly trained ultrasound operators in low resource settings.
Supervised Transfer Learning at Scale for Medical Imaging
Jan 21, 2021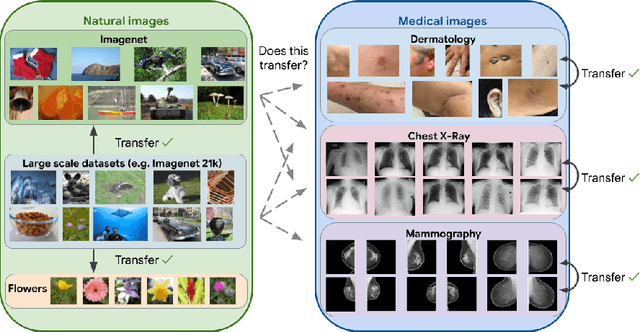
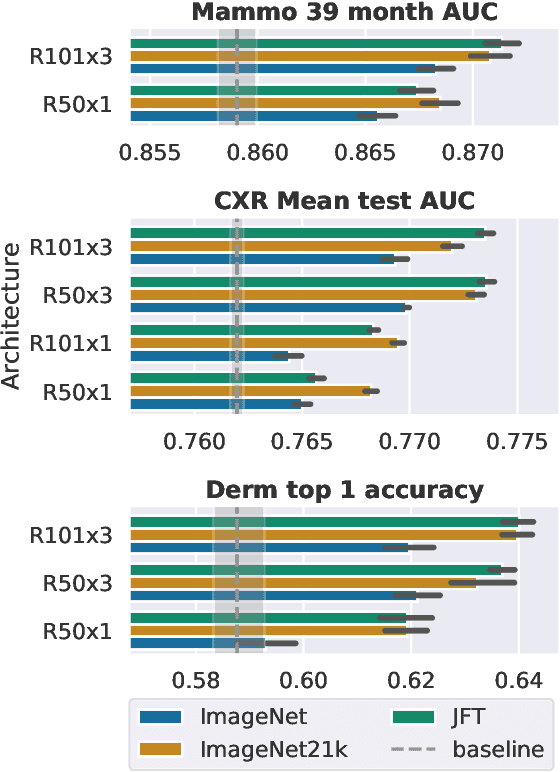
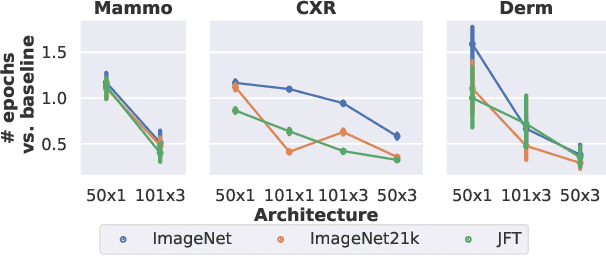
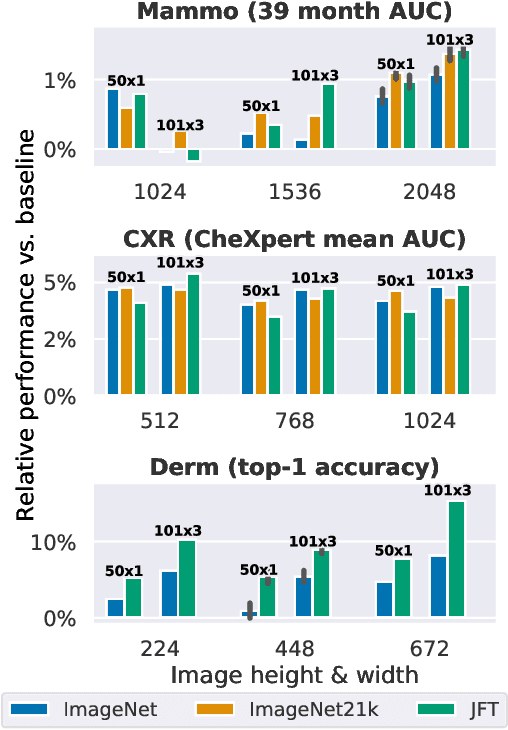
Abstract:Transfer learning is a standard technique to improve performance on tasks with limited data. However, for medical imaging, the value of transfer learning is less clear. This is likely due to the large domain mismatch between the usual natural-image pre-training (e.g. ImageNet) and medical images. However, recent advances in transfer learning have shown substantial improvements from scale. We investigate whether modern methods can change the fortune of transfer learning for medical imaging. For this, we study the class of large-scale pre-trained networks presented by Kolesnikov et al. on three diverse imaging tasks: chest radiography, mammography, and dermatology. We study both transfer performance and critical properties for the deployment in the medical domain, including: out-of-distribution generalization, data-efficiency, sub-group fairness, and uncertainty estimation. Interestingly, we find that for some of these properties transfer from natural to medical images is indeed extremely effective, but only when performed at sufficient scale.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge