Akib Uddin
Discovering novel systemic biomarkers in photos of the external eye
Jul 19, 2022
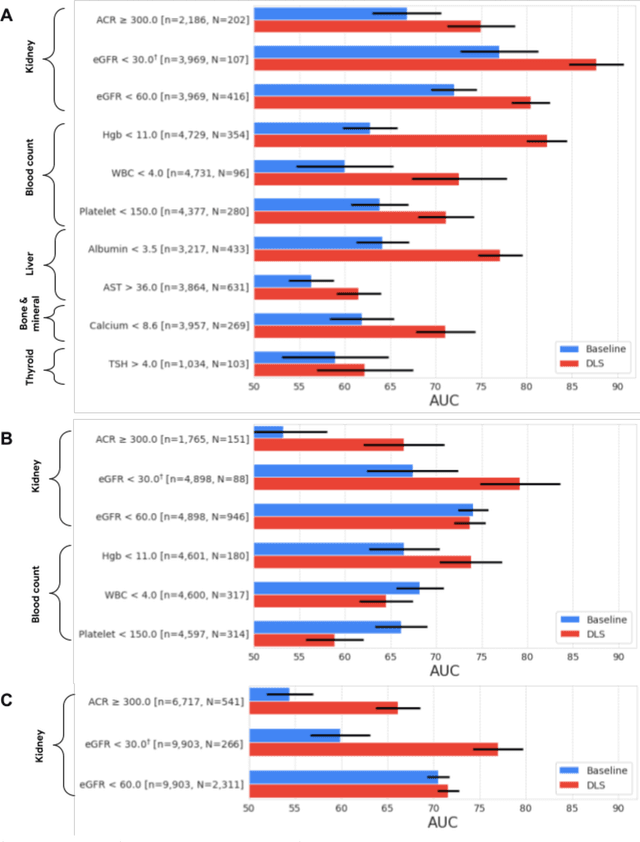
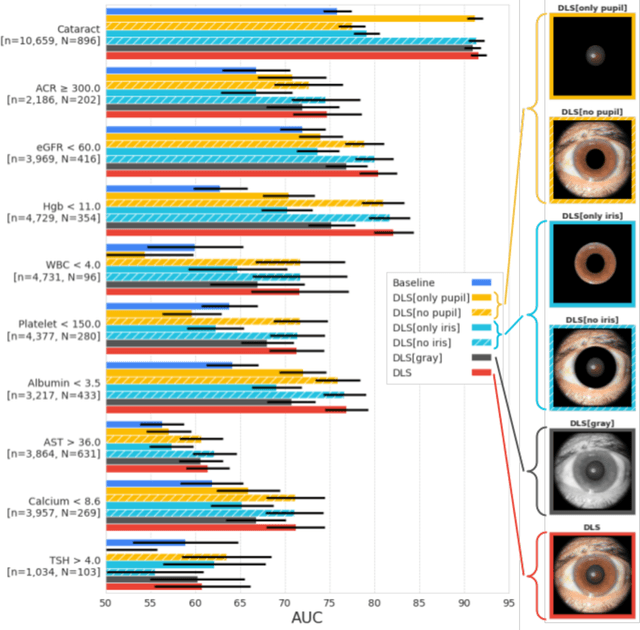
Abstract:External eye photos were recently shown to reveal signs of diabetic retinal disease and elevated HbA1c. In this paper, we evaluate if external eye photos contain information about additional systemic medical conditions. We developed a deep learning system (DLS) that takes external eye photos as input and predicts multiple systemic parameters, such as those related to the liver (albumin, AST); kidney (eGFR estimated using the race-free 2021 CKD-EPI creatinine equation, the urine ACR); bone & mineral (calcium); thyroid (TSH); and blood count (Hgb, WBC, platelets). Development leveraged 151,237 images from 49,015 patients with diabetes undergoing diabetic eye screening in 11 sites across Los Angeles county, CA. Evaluation focused on 9 pre-specified systemic parameters and leveraged 3 validation sets (A, B, C) spanning 28,869 patients with and without diabetes undergoing eye screening in 3 independent sites in Los Angeles County, CA, and the greater Atlanta area, GA. We compared against baseline models incorporating available clinicodemographic variables (e.g. age, sex, race/ethnicity, years with diabetes). Relative to the baseline, the DLS achieved statistically significant superior performance at detecting AST>36, calcium<8.6, eGFR<60, Hgb<11, platelets<150, ACR>=300, and WBC<4 on validation set A (a patient population similar to the development sets), where the AUC of DLS exceeded that of the baseline by 5.2-19.4%. On validation sets B and C, with substantial patient population differences compared to the development sets, the DLS outperformed the baseline for ACR>=300 and Hgb<11 by 7.3-13.2%. Our findings provide further evidence that external eye photos contain important biomarkers of systemic health spanning multiple organ systems. Further work is needed to investigate whether and how these biomarkers can be translated into clinical impact.
Enabling faster and more reliable sonographic assessment of gestational age through machine learning
Mar 22, 2022



Abstract:Fetal ultrasounds are an essential part of prenatal care and can be used to estimate gestational age (GA). Accurate GA assessment is important for providing appropriate prenatal care throughout pregnancy and identifying complications such as fetal growth disorders. Since derivation of GA from manual fetal biometry measurements (head, abdomen, femur) are operator-dependent and time-consuming, there have been a number of research efforts focused on using artificial intelligence (AI) models to estimate GA using standard biometry images, but there is still room to improve the accuracy and reliability of these AI systems for widescale adoption. To improve GA estimates, without significant change to provider workflows, we leverage AI to interpret standard plane ultrasound images as well as 'fly-to' ultrasound videos, which are 5-10s videos automatically recorded as part of the standard of care before the still image is captured. We developed and validated three AI models: an image model using standard plane images, a video model using fly-to videos, and an ensemble model (combining both image and video). All three were statistically superior to standard fetal biometry-based GA estimates derived by expert sonographers, the ensemble model has the lowest mean absolute error (MAE) compared to the clinical standard fetal biometry (mean difference: -1.51 $\pm$ 3.96 days, 95% CI [-1.9, -1.1]) on a test set that consisted of 404 participants. We showed that our models outperform standard biometry by a more substantial margin on fetuses that were small for GA. Our AI models have the potential to empower trained operators to estimate GA with higher accuracy while reducing the amount of time required and user variability in measurement acquisition.
AI system for fetal ultrasound in low-resource settings
Mar 18, 2022



Abstract:Despite considerable progress in maternal healthcare, maternal and perinatal deaths remain high in low-to-middle income countries. Fetal ultrasound is an important component of antenatal care, but shortage of adequately trained healthcare workers has limited its adoption. We developed and validated an artificial intelligence (AI) system that uses novice-acquired "blind sweep" ultrasound videos to estimate gestational age (GA) and fetal malpresentation. We further addressed obstacles that may be encountered in low-resourced settings. Using a simplified sweep protocol with real-time AI feedback on sweep quality, we have demonstrated the generalization of model performance to minimally trained novice ultrasound operators using low cost ultrasound devices with on-device AI integration. The GA model was non-inferior to standard fetal biometry estimates with as few as two sweeps, and the fetal malpresentation model had high AUC-ROCs across operators and devices. Our AI models have the potential to assist in upleveling the capabilities of lightly trained ultrasound operators in low resource settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge