Preeti Singh
A Toolbox for Surfacing Health Equity Harms and Biases in Large Language Models
Mar 18, 2024Abstract:Large language models (LLMs) hold immense promise to serve complex health information needs but also have the potential to introduce harm and exacerbate health disparities. Reliably evaluating equity-related model failures is a critical step toward developing systems that promote health equity. In this work, we present resources and methodologies for surfacing biases with potential to precipitate equity-related harms in long-form, LLM-generated answers to medical questions and then conduct an empirical case study with Med-PaLM 2, resulting in the largest human evaluation study in this area to date. Our contributions include a multifactorial framework for human assessment of LLM-generated answers for biases, and EquityMedQA, a collection of seven newly-released datasets comprising both manually-curated and LLM-generated questions enriched for adversarial queries. Both our human assessment framework and dataset design process are grounded in an iterative participatory approach and review of possible biases in Med-PaLM 2 answers to adversarial queries. Through our empirical study, we find that the use of a collection of datasets curated through a variety of methodologies, coupled with a thorough evaluation protocol that leverages multiple assessment rubric designs and diverse rater groups, surfaces biases that may be missed via narrower evaluation approaches. Our experience underscores the importance of using diverse assessment methodologies and involving raters of varying backgrounds and expertise. We emphasize that while our framework can identify specific forms of bias, it is not sufficient to holistically assess whether the deployment of an AI system promotes equitable health outcomes. We hope the broader community leverages and builds on these tools and methods towards realizing a shared goal of LLMs that promote accessible and equitable healthcare for all.
Closing the AI generalization gap by adjusting for dermatology condition distribution differences across clinical settings
Feb 23, 2024Abstract:Recently, there has been great progress in the ability of artificial intelligence (AI) algorithms to classify dermatological conditions from clinical photographs. However, little is known about the robustness of these algorithms in real-world settings where several factors can lead to a loss of generalizability. Understanding and overcoming these limitations will permit the development of generalizable AI that can aid in the diagnosis of skin conditions across a variety of clinical settings. In this retrospective study, we demonstrate that differences in skin condition distribution, rather than in demographics or image capture mode are the main source of errors when an AI algorithm is evaluated on data from a previously unseen source. We demonstrate a series of steps to close this generalization gap, requiring progressively more information about the new source, ranging from the condition distribution to training data enriched for data less frequently seen during training. Our results also suggest comparable performance from end-to-end fine tuning versus fine tuning solely the classification layer on top of a frozen embedding model. Our approach can inform the adaptation of AI algorithms to new settings, based on the information and resources available.
ELIXR: Towards a general purpose X-ray artificial intelligence system through alignment of large language models and radiology vision encoders
Aug 02, 2023Abstract:Our approach, which we call Embeddings for Language/Image-aligned X-Rays, or ELIXR, leverages a language-aligned image encoder combined or grafted onto a fixed LLM, PaLM 2, to perform a broad range of tasks. We train this lightweight adapter architecture using images paired with corresponding free-text radiology reports from the MIMIC-CXR dataset. ELIXR achieved state-of-the-art performance on zero-shot chest X-ray (CXR) classification (mean AUC of 0.850 across 13 findings), data-efficient CXR classification (mean AUCs of 0.893 and 0.898 across five findings (atelectasis, cardiomegaly, consolidation, pleural effusion, and pulmonary edema) for 1% (~2,200 images) and 10% (~22,000 images) training data), and semantic search (0.76 normalized discounted cumulative gain (NDCG) across nineteen queries, including perfect retrieval on twelve of them). Compared to existing data-efficient methods including supervised contrastive learning (SupCon), ELIXR required two orders of magnitude less data to reach similar performance. ELIXR also showed promise on CXR vision-language tasks, demonstrating overall accuracies of 58.7% and 62.5% on visual question answering and report quality assurance tasks, respectively. These results suggest that ELIXR is a robust and versatile approach to CXR AI.
Discovering novel systemic biomarkers in photos of the external eye
Jul 19, 2022
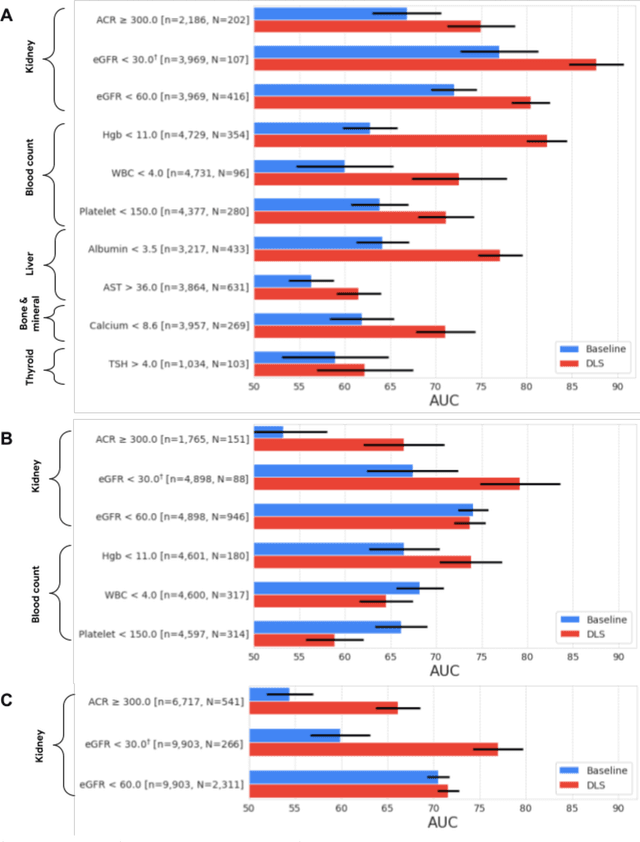
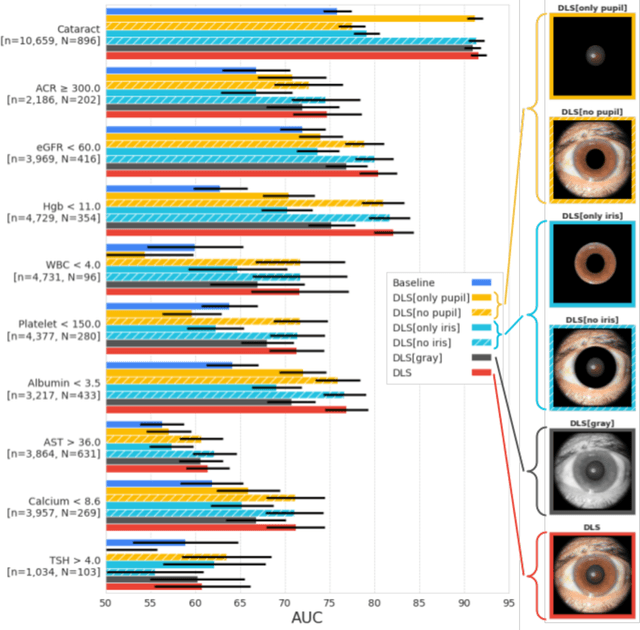
Abstract:External eye photos were recently shown to reveal signs of diabetic retinal disease and elevated HbA1c. In this paper, we evaluate if external eye photos contain information about additional systemic medical conditions. We developed a deep learning system (DLS) that takes external eye photos as input and predicts multiple systemic parameters, such as those related to the liver (albumin, AST); kidney (eGFR estimated using the race-free 2021 CKD-EPI creatinine equation, the urine ACR); bone & mineral (calcium); thyroid (TSH); and blood count (Hgb, WBC, platelets). Development leveraged 151,237 images from 49,015 patients with diabetes undergoing diabetic eye screening in 11 sites across Los Angeles county, CA. Evaluation focused on 9 pre-specified systemic parameters and leveraged 3 validation sets (A, B, C) spanning 28,869 patients with and without diabetes undergoing eye screening in 3 independent sites in Los Angeles County, CA, and the greater Atlanta area, GA. We compared against baseline models incorporating available clinicodemographic variables (e.g. age, sex, race/ethnicity, years with diabetes). Relative to the baseline, the DLS achieved statistically significant superior performance at detecting AST>36, calcium<8.6, eGFR<60, Hgb<11, platelets<150, ACR>=300, and WBC<4 on validation set A (a patient population similar to the development sets), where the AUC of DLS exceeded that of the baseline by 5.2-19.4%. On validation sets B and C, with substantial patient population differences compared to the development sets, the DLS outperformed the baseline for ACR>=300 and Hgb<11 by 7.3-13.2%. Our findings provide further evidence that external eye photos contain important biomarkers of systemic health spanning multiple organ systems. Further work is needed to investigate whether and how these biomarkers can be translated into clinical impact.
Detecting hidden signs of diabetes in external eye photographs
Nov 23, 2020



Abstract:Diabetes-related retinal conditions can be detected by examining the posterior of the eye. By contrast, examining the anterior of the eye can reveal conditions affecting the front of the eye, such as changes to the eyelids, cornea, or crystalline lens. In this work, we studied whether external photographs of the front of the eye can reveal insights into both diabetic retinal diseases and blood glucose control. We developed a deep learning system (DLS) using external eye photographs of 145,832 patients with diabetes from 301 diabetic retinopathy (DR) screening sites in one US state, and evaluated the DLS on three validation sets containing images from 198 sites in 18 other US states. In validation set A (n=27,415 patients, all undilated), the DLS detected poor blood glucose control (HbA1c > 9%) with an area under receiver operating characteristic curve (AUC) of 70.2; moderate-or-worse DR with an AUC of 75.3; diabetic macular edema with an AUC of 78.0; and vision-threatening DR with an AUC of 79.4. For all 4 prediction tasks, the DLS's AUC was higher (p<0.001) than using available self-reported baseline characteristics (age, sex, race/ethnicity, years with diabetes). In terms of positive predictive value, the predicted top 5% of patients had a 67% chance of having HbA1c > 9%, and a 20% chance of having vision threatening diabetic retinopathy. The results generalized to dilated pupils (validation set B, 5,058 patients) and to a different screening service (validation set C, 10,402 patients). Our results indicate that external eye photographs contain information useful for healthcare providers managing patients with diabetes, and may help prioritize patients for in-person screening. Further work is needed to validate these findings on different devices and patient populations (those without diabetes) to evaluate its utility for remote diagnosis and management.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge