Nenad Tomasev
Advancing Conversational Diagnostic AI with Multimodal Reasoning
May 06, 2025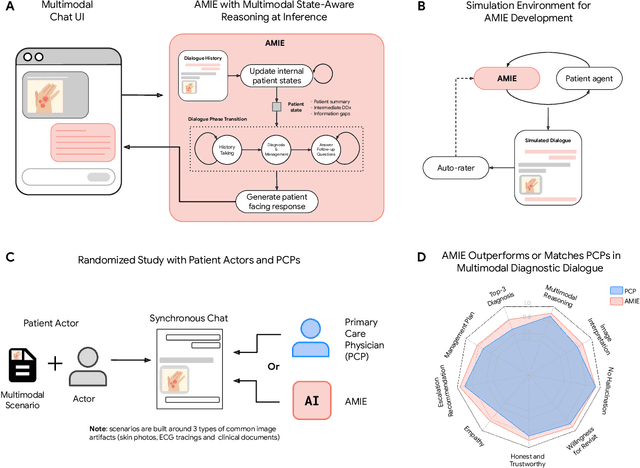
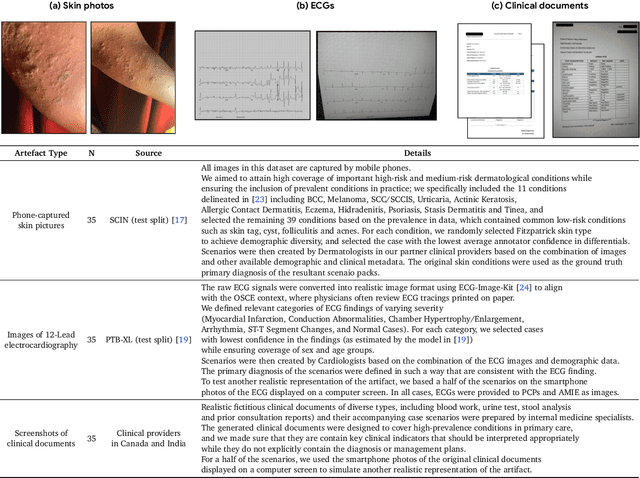
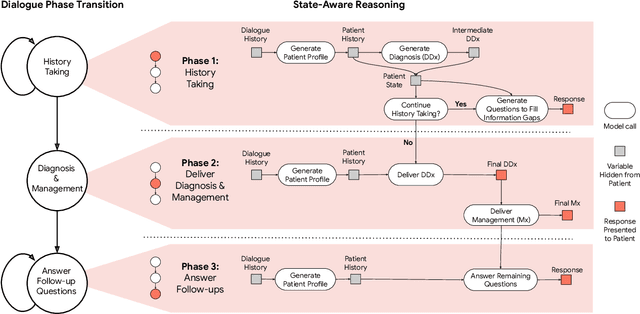
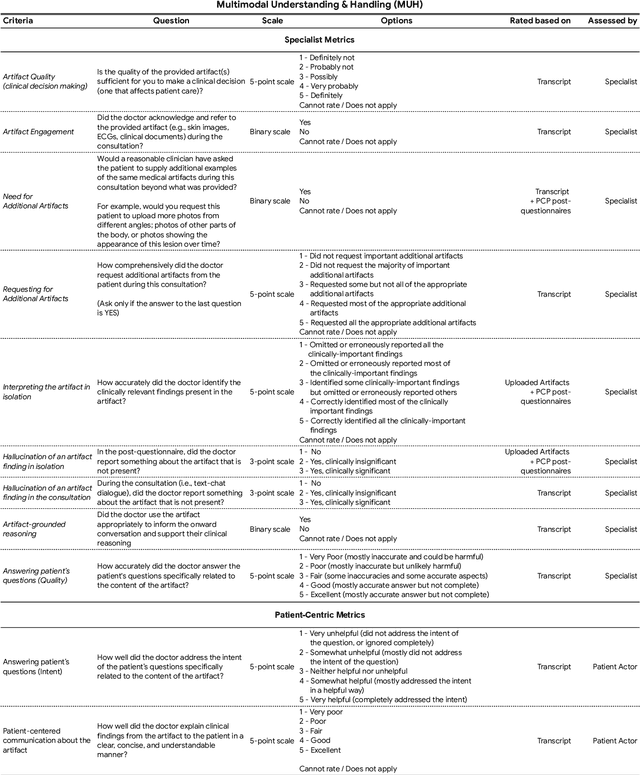
Abstract:Large Language Models (LLMs) have demonstrated great potential for conducting diagnostic conversations but evaluation has been largely limited to language-only interactions, deviating from the real-world requirements of remote care delivery. Instant messaging platforms permit clinicians and patients to upload and discuss multimodal medical artifacts seamlessly in medical consultation, but the ability of LLMs to reason over such data while preserving other attributes of competent diagnostic conversation remains unknown. Here we advance the conversational diagnosis and management performance of the Articulate Medical Intelligence Explorer (AMIE) through a new capability to gather and interpret multimodal data, and reason about this precisely during consultations. Leveraging Gemini 2.0 Flash, our system implements a state-aware dialogue framework, where conversation flow is dynamically controlled by intermediate model outputs reflecting patient states and evolving diagnoses. Follow-up questions are strategically directed by uncertainty in such patient states, leading to a more structured multimodal history-taking process that emulates experienced clinicians. We compared AMIE to primary care physicians (PCPs) in a randomized, blinded, OSCE-style study of chat-based consultations with patient actors. We constructed 105 evaluation scenarios using artifacts like smartphone skin photos, ECGs, and PDFs of clinical documents across diverse conditions and demographics. Our rubric assessed multimodal capabilities and other clinically meaningful axes like history-taking, diagnostic accuracy, management reasoning, communication, and empathy. Specialist evaluation showed AMIE to be superior to PCPs on 7/9 multimodal and 29/32 non-multimodal axes (including diagnostic accuracy). The results show clear progress in multimodal conversational diagnostic AI, but real-world translation needs further research.
Towards an AI co-scientist
Feb 26, 2025Abstract:Scientific discovery relies on scientists generating novel hypotheses that undergo rigorous experimental validation. To augment this process, we introduce an AI co-scientist, a multi-agent system built on Gemini 2.0. The AI co-scientist is intended to help uncover new, original knowledge and to formulate demonstrably novel research hypotheses and proposals, building upon prior evidence and aligned to scientist-provided research objectives and guidance. The system's design incorporates a generate, debate, and evolve approach to hypothesis generation, inspired by the scientific method and accelerated by scaling test-time compute. Key contributions include: (1) a multi-agent architecture with an asynchronous task execution framework for flexible compute scaling; (2) a tournament evolution process for self-improving hypotheses generation. Automated evaluations show continued benefits of test-time compute, improving hypothesis quality. While general purpose, we focus development and validation in three biomedical areas: drug repurposing, novel target discovery, and explaining mechanisms of bacterial evolution and anti-microbial resistance. For drug repurposing, the system proposes candidates with promising validation findings, including candidates for acute myeloid leukemia that show tumor inhibition in vitro at clinically applicable concentrations. For novel target discovery, the AI co-scientist proposed new epigenetic targets for liver fibrosis, validated by anti-fibrotic activity and liver cell regeneration in human hepatic organoids. Finally, the AI co-scientist recapitulated unpublished experimental results via a parallel in silico discovery of a novel gene transfer mechanism in bacterial evolution. These results, detailed in separate, co-timed reports, demonstrate the potential to augment biomedical and scientific discovery and usher an era of AI empowered scientists.
Contextual Evaluation of Large Language Models for Classifying Tropical and Infectious Diseases
Sep 13, 2024Abstract:While large language models (LLMs) have shown promise for medical question answering, there is limited work focused on tropical and infectious disease-specific exploration. We build on an opensource tropical and infectious diseases (TRINDs) dataset, expanding it to include demographic and semantic clinical and consumer augmentations yielding 11000+ prompts. We evaluate LLM performance on these, comparing generalist and medical LLMs, as well as LLM outcomes to human experts. We demonstrate through systematic experimentation, the benefit of contextual information such as demographics, location, gender, risk factors for optimal LLM response. Finally we develop a prototype of TRINDs-LM, a research tool that provides a playground to navigate how context impacts LLM outputs for health.
Capabilities of Gemini Models in Medicine
May 01, 2024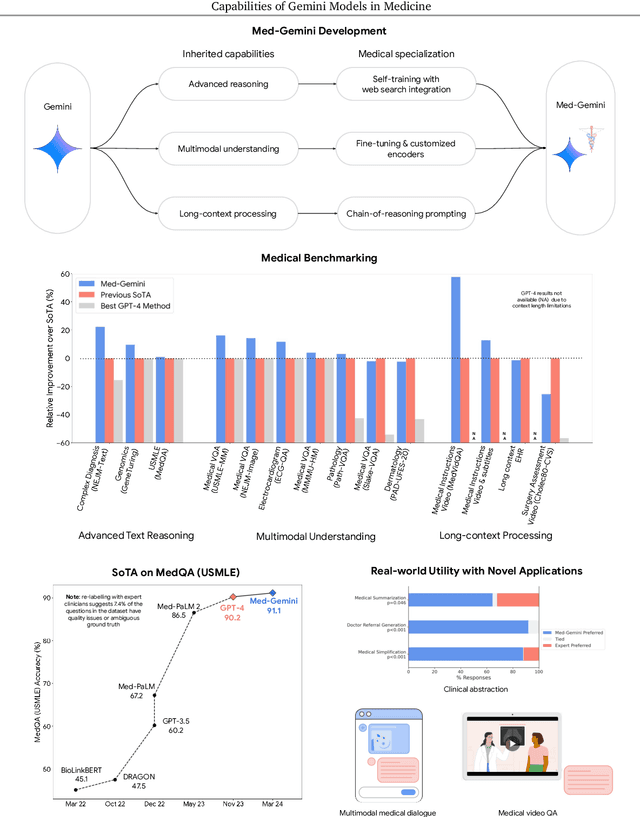
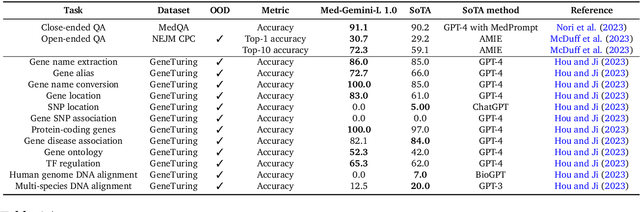
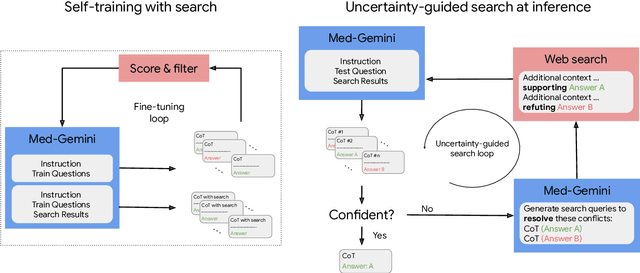
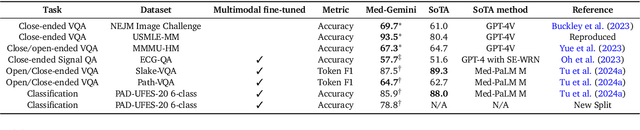
Abstract:Excellence in a wide variety of medical applications poses considerable challenges for AI, requiring advanced reasoning, access to up-to-date medical knowledge and understanding of complex multimodal data. Gemini models, with strong general capabilities in multimodal and long-context reasoning, offer exciting possibilities in medicine. Building on these core strengths of Gemini, we introduce Med-Gemini, a family of highly capable multimodal models that are specialized in medicine with the ability to seamlessly use web search, and that can be efficiently tailored to novel modalities using custom encoders. We evaluate Med-Gemini on 14 medical benchmarks, establishing new state-of-the-art (SoTA) performance on 10 of them, and surpass the GPT-4 model family on every benchmark where a direct comparison is viable, often by a wide margin. On the popular MedQA (USMLE) benchmark, our best-performing Med-Gemini model achieves SoTA performance of 91.1% accuracy, using a novel uncertainty-guided search strategy. On 7 multimodal benchmarks including NEJM Image Challenges and MMMU (health & medicine), Med-Gemini improves over GPT-4V by an average relative margin of 44.5%. We demonstrate the effectiveness of Med-Gemini's long-context capabilities through SoTA performance on a needle-in-a-haystack retrieval task from long de-identified health records and medical video question answering, surpassing prior bespoke methods using only in-context learning. Finally, Med-Gemini's performance suggests real-world utility by surpassing human experts on tasks such as medical text summarization, alongside demonstrations of promising potential for multimodal medical dialogue, medical research and education. Taken together, our results offer compelling evidence for Med-Gemini's potential, although further rigorous evaluation will be crucial before real-world deployment in this safety-critical domain.
A Toolbox for Surfacing Health Equity Harms and Biases in Large Language Models
Mar 18, 2024Abstract:Large language models (LLMs) hold immense promise to serve complex health information needs but also have the potential to introduce harm and exacerbate health disparities. Reliably evaluating equity-related model failures is a critical step toward developing systems that promote health equity. In this work, we present resources and methodologies for surfacing biases with potential to precipitate equity-related harms in long-form, LLM-generated answers to medical questions and then conduct an empirical case study with Med-PaLM 2, resulting in the largest human evaluation study in this area to date. Our contributions include a multifactorial framework for human assessment of LLM-generated answers for biases, and EquityMedQA, a collection of seven newly-released datasets comprising both manually-curated and LLM-generated questions enriched for adversarial queries. Both our human assessment framework and dataset design process are grounded in an iterative participatory approach and review of possible biases in Med-PaLM 2 answers to adversarial queries. Through our empirical study, we find that the use of a collection of datasets curated through a variety of methodologies, coupled with a thorough evaluation protocol that leverages multiple assessment rubric designs and diverse rater groups, surfaces biases that may be missed via narrower evaluation approaches. Our experience underscores the importance of using diverse assessment methodologies and involving raters of varying backgrounds and expertise. We emphasize that while our framework can identify specific forms of bias, it is not sufficient to holistically assess whether the deployment of an AI system promotes equitable health outcomes. We hope the broader community leverages and builds on these tools and methods towards realizing a shared goal of LLMs that promote accessible and equitable healthcare for all.
Towards Conversational Diagnostic AI
Jan 11, 2024Abstract:At the heart of medicine lies the physician-patient dialogue, where skillful history-taking paves the way for accurate diagnosis, effective management, and enduring trust. Artificial Intelligence (AI) systems capable of diagnostic dialogue could increase accessibility, consistency, and quality of care. However, approximating clinicians' expertise is an outstanding grand challenge. Here, we introduce AMIE (Articulate Medical Intelligence Explorer), a Large Language Model (LLM) based AI system optimized for diagnostic dialogue. AMIE uses a novel self-play based simulated environment with automated feedback mechanisms for scaling learning across diverse disease conditions, specialties, and contexts. We designed a framework for evaluating clinically-meaningful axes of performance including history-taking, diagnostic accuracy, management reasoning, communication skills, and empathy. We compared AMIE's performance to that of primary care physicians (PCPs) in a randomized, double-blind crossover study of text-based consultations with validated patient actors in the style of an Objective Structured Clinical Examination (OSCE). The study included 149 case scenarios from clinical providers in Canada, the UK, and India, 20 PCPs for comparison with AMIE, and evaluations by specialist physicians and patient actors. AMIE demonstrated greater diagnostic accuracy and superior performance on 28 of 32 axes according to specialist physicians and 24 of 26 axes according to patient actors. Our research has several limitations and should be interpreted with appropriate caution. Clinicians were limited to unfamiliar synchronous text-chat which permits large-scale LLM-patient interactions but is not representative of usual clinical practice. While further research is required before AMIE could be translated to real-world settings, the results represent a milestone towards conversational diagnostic AI.
Gemini: A Family of Highly Capable Multimodal Models
Dec 19, 2023Abstract:This report introduces a new family of multimodal models, Gemini, that exhibit remarkable capabilities across image, audio, video, and text understanding. The Gemini family consists of Ultra, Pro, and Nano sizes, suitable for applications ranging from complex reasoning tasks to on-device memory-constrained use-cases. Evaluation on a broad range of benchmarks shows that our most-capable Gemini Ultra model advances the state of the art in 30 of 32 of these benchmarks - notably being the first model to achieve human-expert performance on the well-studied exam benchmark MMLU, and improving the state of the art in every one of the 20 multimodal benchmarks we examined. We believe that the new capabilities of Gemini models in cross-modal reasoning and language understanding will enable a wide variety of use cases and we discuss our approach toward deploying them responsibly to users.
Bridging the Human-AI Knowledge Gap: Concept Discovery and Transfer in AlphaZero
Oct 25, 2023Abstract:Artificial Intelligence (AI) systems have made remarkable progress, attaining super-human performance across various domains. This presents us with an opportunity to further human knowledge and improve human expert performance by leveraging the hidden knowledge encoded within these highly performant AI systems. Yet, this knowledge is often hard to extract, and may be hard to understand or learn from. Here, we show that this is possible by proposing a new method that allows us to extract new chess concepts in AlphaZero, an AI system that mastered the game of chess via self-play without human supervision. Our analysis indicates that AlphaZero may encode knowledge that extends beyond the existing human knowledge, but knowledge that is ultimately not beyond human grasp, and can be successfully learned from. In a human study, we show that these concepts are learnable by top human experts, as four top chess grandmasters show improvements in solving the presented concept prototype positions. This marks an important first milestone in advancing the frontier of human knowledge by leveraging AI; a development that could bear profound implications and help us shape how we interact with AI systems across many AI applications.
Diversifying AI: Towards Creative Chess with AlphaZero
Aug 29, 2023



Abstract:In recent years, Artificial Intelligence (AI) systems have surpassed human intelligence in a variety of computational tasks. However, AI systems, like humans, make mistakes, have blind spots, hallucinate, and struggle to generalize to new situations. This work explores whether AI can benefit from creative decision-making mechanisms when pushed to the limits of its computational rationality. In particular, we investigate whether a team of diverse AI systems can outperform a single AI in challenging tasks by generating more ideas as a group and then selecting the best ones. We study this question in the game of chess, the so-called drosophila of AI. We build on AlphaZero (AZ) and extend it to represent a league of agents via a latent-conditioned architecture, which we call AZ_db. We train AZ_db to generate a wider range of ideas using behavioral diversity techniques and select the most promising ones with sub-additive planning. Our experiments suggest that AZ_db plays chess in diverse ways, solves more puzzles as a group and outperforms a more homogeneous team. Notably, AZ_db solves twice as many challenging puzzles as AZ, including the challenging Penrose positions. When playing chess from different openings, we notice that players in AZ_db specialize in different openings, and that selecting a player for each opening using sub-additive planning results in a 50 Elo improvement over AZ. Our findings suggest that diversity bonuses emerge in teams of AI agents, just as they do in teams of humans and that diversity is a valuable asset in solving computationally hard problems.
Towards Expert-Level Medical Question Answering with Large Language Models
May 16, 2023



Abstract:Recent artificial intelligence (AI) systems have reached milestones in "grand challenges" ranging from Go to protein-folding. The capability to retrieve medical knowledge, reason over it, and answer medical questions comparably to physicians has long been viewed as one such grand challenge. Large language models (LLMs) have catalyzed significant progress in medical question answering; Med-PaLM was the first model to exceed a "passing" score in US Medical Licensing Examination (USMLE) style questions with a score of 67.2% on the MedQA dataset. However, this and other prior work suggested significant room for improvement, especially when models' answers were compared to clinicians' answers. Here we present Med-PaLM 2, which bridges these gaps by leveraging a combination of base LLM improvements (PaLM 2), medical domain finetuning, and prompting strategies including a novel ensemble refinement approach. Med-PaLM 2 scored up to 86.5% on the MedQA dataset, improving upon Med-PaLM by over 19% and setting a new state-of-the-art. We also observed performance approaching or exceeding state-of-the-art across MedMCQA, PubMedQA, and MMLU clinical topics datasets. We performed detailed human evaluations on long-form questions along multiple axes relevant to clinical applications. In pairwise comparative ranking of 1066 consumer medical questions, physicians preferred Med-PaLM 2 answers to those produced by physicians on eight of nine axes pertaining to clinical utility (p < 0.001). We also observed significant improvements compared to Med-PaLM on every evaluation axis (p < 0.001) on newly introduced datasets of 240 long-form "adversarial" questions to probe LLM limitations. While further studies are necessary to validate the efficacy of these models in real-world settings, these results highlight rapid progress towards physician-level performance in medical question answering.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge