Anil Palepu
Advancing Conversational Diagnostic AI with Multimodal Reasoning
May 06, 2025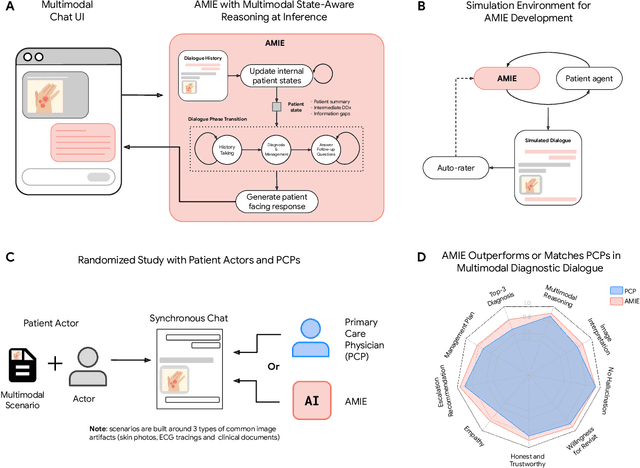
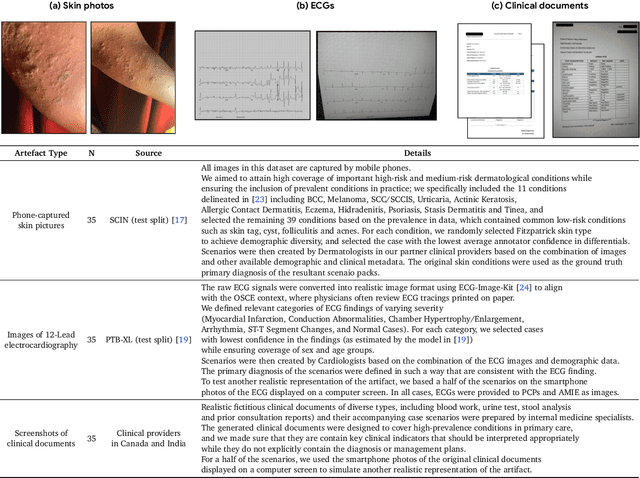
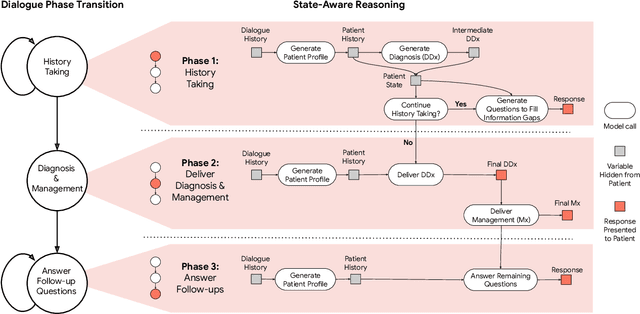
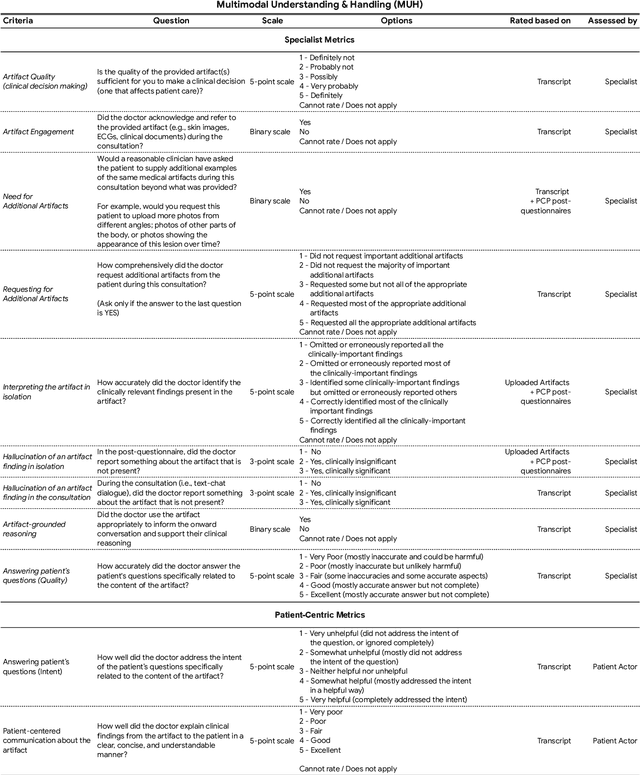
Abstract:Large Language Models (LLMs) have demonstrated great potential for conducting diagnostic conversations but evaluation has been largely limited to language-only interactions, deviating from the real-world requirements of remote care delivery. Instant messaging platforms permit clinicians and patients to upload and discuss multimodal medical artifacts seamlessly in medical consultation, but the ability of LLMs to reason over such data while preserving other attributes of competent diagnostic conversation remains unknown. Here we advance the conversational diagnosis and management performance of the Articulate Medical Intelligence Explorer (AMIE) through a new capability to gather and interpret multimodal data, and reason about this precisely during consultations. Leveraging Gemini 2.0 Flash, our system implements a state-aware dialogue framework, where conversation flow is dynamically controlled by intermediate model outputs reflecting patient states and evolving diagnoses. Follow-up questions are strategically directed by uncertainty in such patient states, leading to a more structured multimodal history-taking process that emulates experienced clinicians. We compared AMIE to primary care physicians (PCPs) in a randomized, blinded, OSCE-style study of chat-based consultations with patient actors. We constructed 105 evaluation scenarios using artifacts like smartphone skin photos, ECGs, and PDFs of clinical documents across diverse conditions and demographics. Our rubric assessed multimodal capabilities and other clinically meaningful axes like history-taking, diagnostic accuracy, management reasoning, communication, and empathy. Specialist evaluation showed AMIE to be superior to PCPs on 7/9 multimodal and 29/32 non-multimodal axes (including diagnostic accuracy). The results show clear progress in multimodal conversational diagnostic AI, but real-world translation needs further research.
Towards Conversational AI for Disease Management
Mar 08, 2025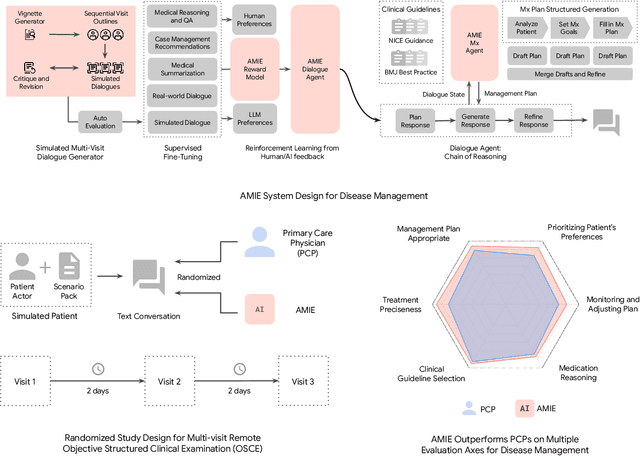

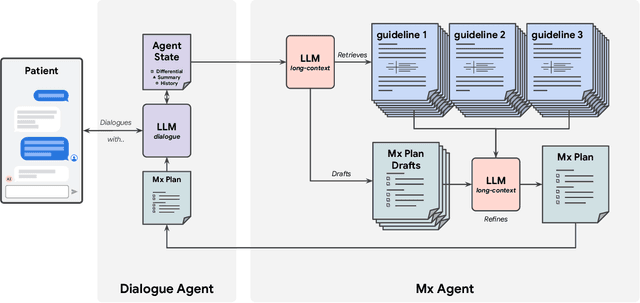
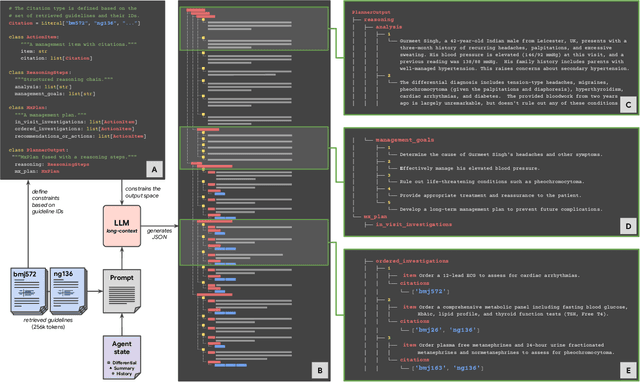
Abstract:While large language models (LLMs) have shown promise in diagnostic dialogue, their capabilities for effective management reasoning - including disease progression, therapeutic response, and safe medication prescription - remain under-explored. We advance the previously demonstrated diagnostic capabilities of the Articulate Medical Intelligence Explorer (AMIE) through a new LLM-based agentic system optimised for clinical management and dialogue, incorporating reasoning over the evolution of disease and multiple patient visit encounters, response to therapy, and professional competence in medication prescription. To ground its reasoning in authoritative clinical knowledge, AMIE leverages Gemini's long-context capabilities, combining in-context retrieval with structured reasoning to align its output with relevant and up-to-date clinical practice guidelines and drug formularies. In a randomized, blinded virtual Objective Structured Clinical Examination (OSCE) study, AMIE was compared to 21 primary care physicians (PCPs) across 100 multi-visit case scenarios designed to reflect UK NICE Guidance and BMJ Best Practice guidelines. AMIE was non-inferior to PCPs in management reasoning as assessed by specialist physicians and scored better in both preciseness of treatments and investigations, and in its alignment with and grounding of management plans in clinical guidelines. To benchmark medication reasoning, we developed RxQA, a multiple-choice question benchmark derived from two national drug formularies (US, UK) and validated by board-certified pharmacists. While AMIE and PCPs both benefited from the ability to access external drug information, AMIE outperformed PCPs on higher difficulty questions. While further research would be needed before real-world translation, AMIE's strong performance across evaluations marks a significant step towards conversational AI as a tool in disease management.
Towards an AI co-scientist
Feb 26, 2025Abstract:Scientific discovery relies on scientists generating novel hypotheses that undergo rigorous experimental validation. To augment this process, we introduce an AI co-scientist, a multi-agent system built on Gemini 2.0. The AI co-scientist is intended to help uncover new, original knowledge and to formulate demonstrably novel research hypotheses and proposals, building upon prior evidence and aligned to scientist-provided research objectives and guidance. The system's design incorporates a generate, debate, and evolve approach to hypothesis generation, inspired by the scientific method and accelerated by scaling test-time compute. Key contributions include: (1) a multi-agent architecture with an asynchronous task execution framework for flexible compute scaling; (2) a tournament evolution process for self-improving hypotheses generation. Automated evaluations show continued benefits of test-time compute, improving hypothesis quality. While general purpose, we focus development and validation in three biomedical areas: drug repurposing, novel target discovery, and explaining mechanisms of bacterial evolution and anti-microbial resistance. For drug repurposing, the system proposes candidates with promising validation findings, including candidates for acute myeloid leukemia that show tumor inhibition in vitro at clinically applicable concentrations. For novel target discovery, the AI co-scientist proposed new epigenetic targets for liver fibrosis, validated by anti-fibrotic activity and liver cell regeneration in human hepatic organoids. Finally, the AI co-scientist recapitulated unpublished experimental results via a parallel in silico discovery of a novel gene transfer mechanism in bacterial evolution. These results, detailed in separate, co-timed reports, demonstrate the potential to augment biomedical and scientific discovery and usher an era of AI empowered scientists.
Capabilities of Gemini Models in Medicine
May 01, 2024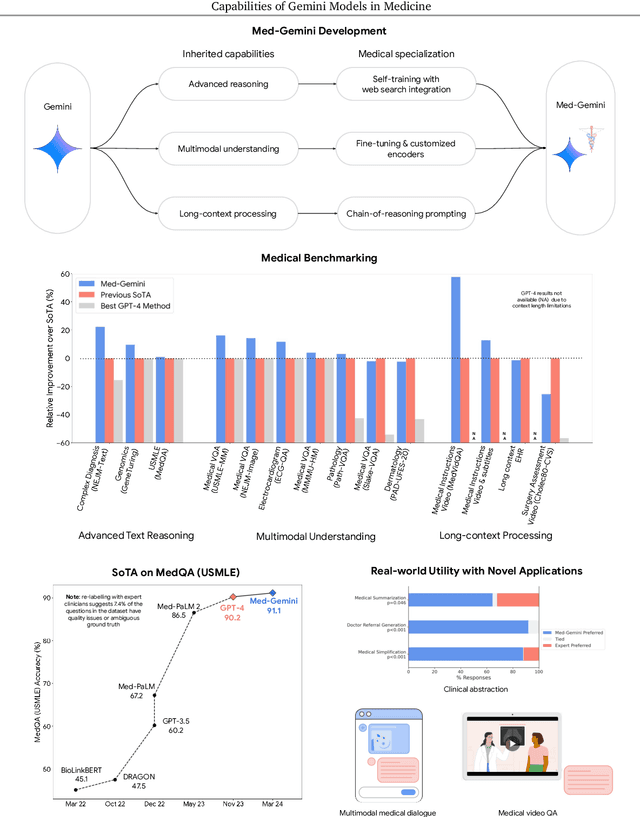
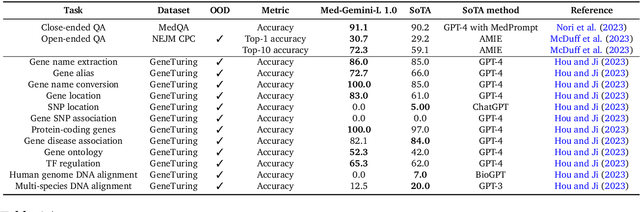
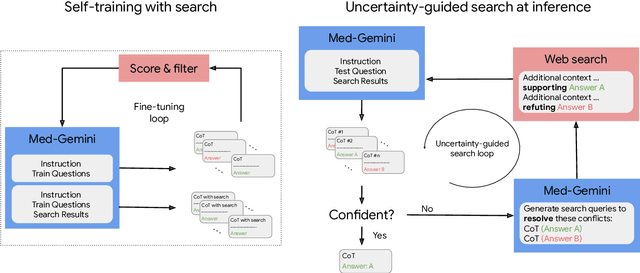
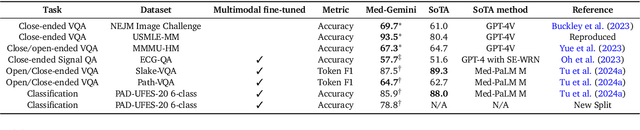
Abstract:Excellence in a wide variety of medical applications poses considerable challenges for AI, requiring advanced reasoning, access to up-to-date medical knowledge and understanding of complex multimodal data. Gemini models, with strong general capabilities in multimodal and long-context reasoning, offer exciting possibilities in medicine. Building on these core strengths of Gemini, we introduce Med-Gemini, a family of highly capable multimodal models that are specialized in medicine with the ability to seamlessly use web search, and that can be efficiently tailored to novel modalities using custom encoders. We evaluate Med-Gemini on 14 medical benchmarks, establishing new state-of-the-art (SoTA) performance on 10 of them, and surpass the GPT-4 model family on every benchmark where a direct comparison is viable, often by a wide margin. On the popular MedQA (USMLE) benchmark, our best-performing Med-Gemini model achieves SoTA performance of 91.1% accuracy, using a novel uncertainty-guided search strategy. On 7 multimodal benchmarks including NEJM Image Challenges and MMMU (health & medicine), Med-Gemini improves over GPT-4V by an average relative margin of 44.5%. We demonstrate the effectiveness of Med-Gemini's long-context capabilities through SoTA performance on a needle-in-a-haystack retrieval task from long de-identified health records and medical video question answering, surpassing prior bespoke methods using only in-context learning. Finally, Med-Gemini's performance suggests real-world utility by surpassing human experts on tasks such as medical text summarization, alongside demonstrations of promising potential for multimodal medical dialogue, medical research and education. Taken together, our results offer compelling evidence for Med-Gemini's potential, although further rigorous evaluation will be crucial before real-world deployment in this safety-critical domain.
Towards Conversational Diagnostic AI
Jan 11, 2024Abstract:At the heart of medicine lies the physician-patient dialogue, where skillful history-taking paves the way for accurate diagnosis, effective management, and enduring trust. Artificial Intelligence (AI) systems capable of diagnostic dialogue could increase accessibility, consistency, and quality of care. However, approximating clinicians' expertise is an outstanding grand challenge. Here, we introduce AMIE (Articulate Medical Intelligence Explorer), a Large Language Model (LLM) based AI system optimized for diagnostic dialogue. AMIE uses a novel self-play based simulated environment with automated feedback mechanisms for scaling learning across diverse disease conditions, specialties, and contexts. We designed a framework for evaluating clinically-meaningful axes of performance including history-taking, diagnostic accuracy, management reasoning, communication skills, and empathy. We compared AMIE's performance to that of primary care physicians (PCPs) in a randomized, double-blind crossover study of text-based consultations with validated patient actors in the style of an Objective Structured Clinical Examination (OSCE). The study included 149 case scenarios from clinical providers in Canada, the UK, and India, 20 PCPs for comparison with AMIE, and evaluations by specialist physicians and patient actors. AMIE demonstrated greater diagnostic accuracy and superior performance on 28 of 32 axes according to specialist physicians and 24 of 26 axes according to patient actors. Our research has several limitations and should be interpreted with appropriate caution. Clinicians were limited to unfamiliar synchronous text-chat which permits large-scale LLM-patient interactions but is not representative of usual clinical practice. While further research is required before AMIE could be translated to real-world settings, the results represent a milestone towards conversational diagnostic AI.
Towards Accurate Differential Diagnosis with Large Language Models
Nov 30, 2023



Abstract:An accurate differential diagnosis (DDx) is a cornerstone of medical care, often reached through an iterative process of interpretation that combines clinical history, physical examination, investigations and procedures. Interactive interfaces powered by Large Language Models (LLMs) present new opportunities to both assist and automate aspects of this process. In this study, we introduce an LLM optimized for diagnostic reasoning, and evaluate its ability to generate a DDx alone or as an aid to clinicians. 20 clinicians evaluated 302 challenging, real-world medical cases sourced from the New England Journal of Medicine (NEJM) case reports. Each case report was read by two clinicians, who were randomized to one of two assistive conditions: either assistance from search engines and standard medical resources, or LLM assistance in addition to these tools. All clinicians provided a baseline, unassisted DDx prior to using the respective assistive tools. Our LLM for DDx exhibited standalone performance that exceeded that of unassisted clinicians (top-10 accuracy 59.1% vs 33.6%, [p = 0.04]). Comparing the two assisted study arms, the DDx quality score was higher for clinicians assisted by our LLM (top-10 accuracy 51.7%) compared to clinicians without its assistance (36.1%) (McNemar's Test: 45.7, p < 0.01) and clinicians with search (44.4%) (4.75, p = 0.03). Further, clinicians assisted by our LLM arrived at more comprehensive differential lists than those without its assistance. Our study suggests that our LLM for DDx has potential to improve clinicians' diagnostic reasoning and accuracy in challenging cases, meriting further real-world evaluation for its ability to empower physicians and widen patients' access to specialist-level expertise.
Conformal Prediction with Large Language Models for Multi-Choice Question Answering
Jun 01, 2023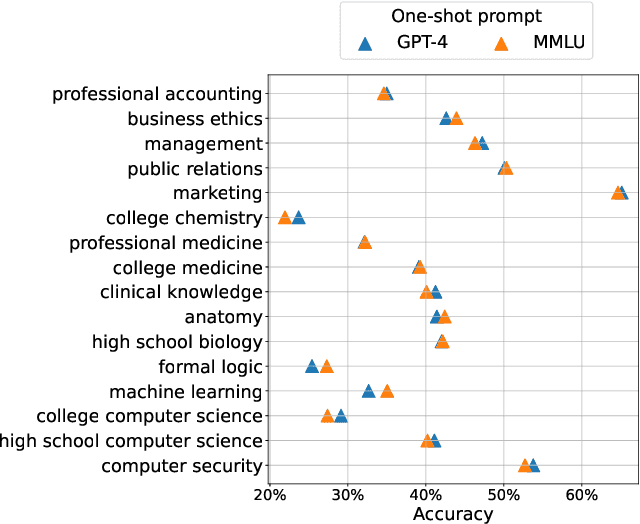
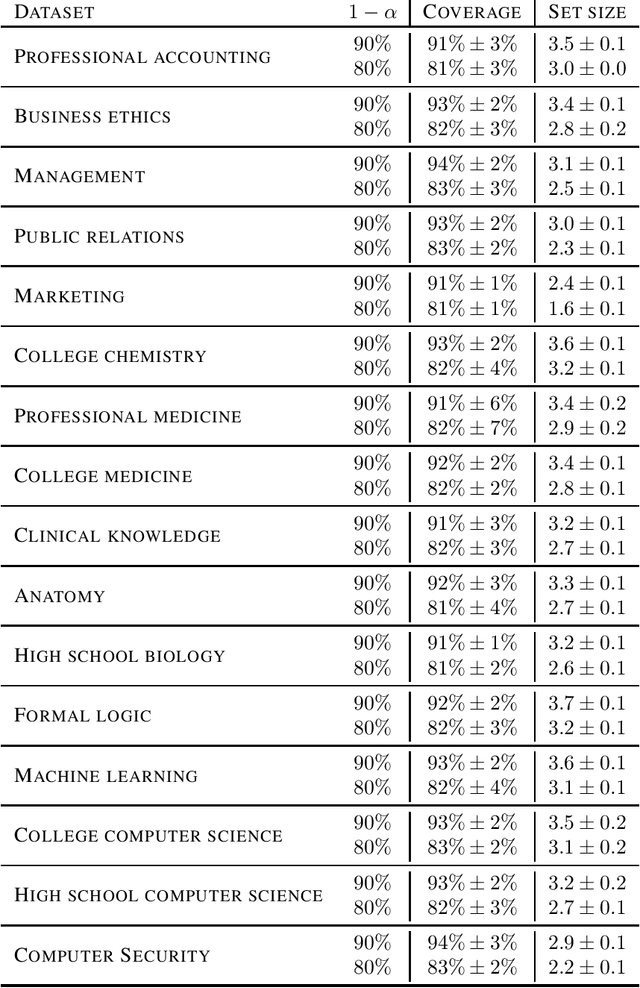
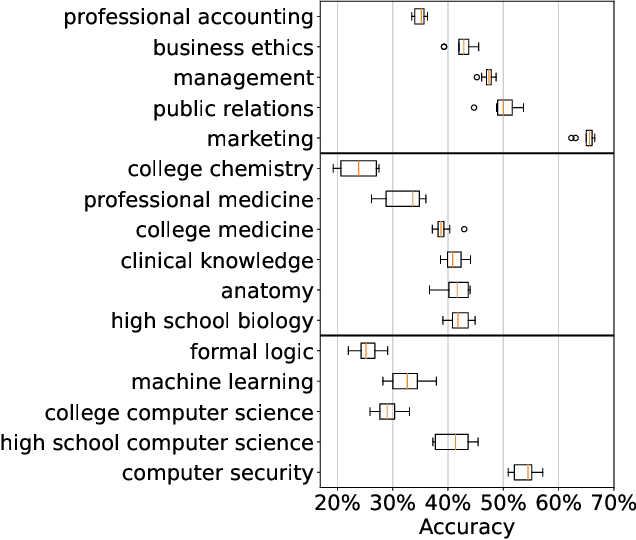
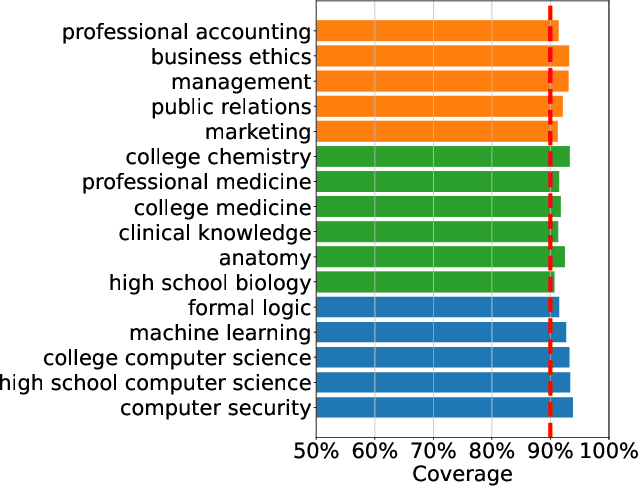
Abstract:As large language models continue to be widely developed, robust uncertainty quantification techniques will become crucial for their safe deployment in high-stakes scenarios. In this work, we explore how conformal prediction can be used to provide uncertainty quantification in language models for the specific task of multiple-choice question-answering. We find that the uncertainty estimates from conformal prediction are tightly correlated with prediction accuracy. This observation can be useful for downstream applications such as selective classification and filtering out low-quality predictions. We also investigate the exchangeability assumption required by conformal prediction to out-of-subject questions, which may be a more realistic scenario for many practical applications. Our work contributes towards more trustworthy and reliable usage of large language models in safety-critical situations, where robust guarantees of error rate are required.
TIER: Text-Image Entropy Regularization for CLIP-style models
Dec 13, 2022Abstract:In this paper, we study the effect of a novel regularization scheme on contrastive language-image pre-trained (CLIP) models. Our approach is based on the observation that, in many domains, text tokens should only describe a small number of image regions and, likewise, each image region should correspond to only a few text tokens. In CLIP-style models, this implies that text-token embeddings should have high similarity to only a small number of image-patch embeddings for a given image-text pair. We formalize this observation using a novel regularization scheme that penalizes the entropy of the text-token to image-patch similarity scores. We qualitatively and quantitatively demonstrate that the proposed regularization scheme shrinks the text-token and image-patch similarity scores towards zero, thus achieving the desired effect. We demonstrate the promise of our approach in an important medical context where this underlying hypothesis naturally arises. Using our proposed approach, we achieve state of the art (SOTA) zero-shot performance on all tasks from the CheXpert chest x-ray dataset, outperforming an unregularized version of the model and several recently published self-supervised models.
Towards Reliable Zero Shot Classification in Self-Supervised Models with Conformal Prediction
Oct 27, 2022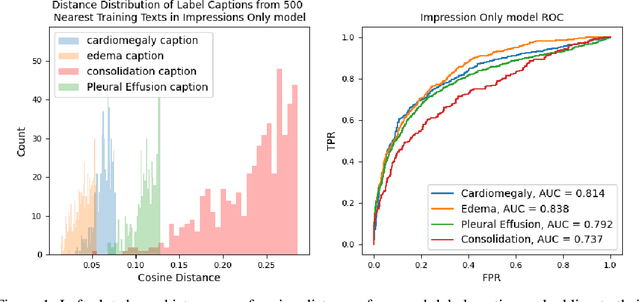
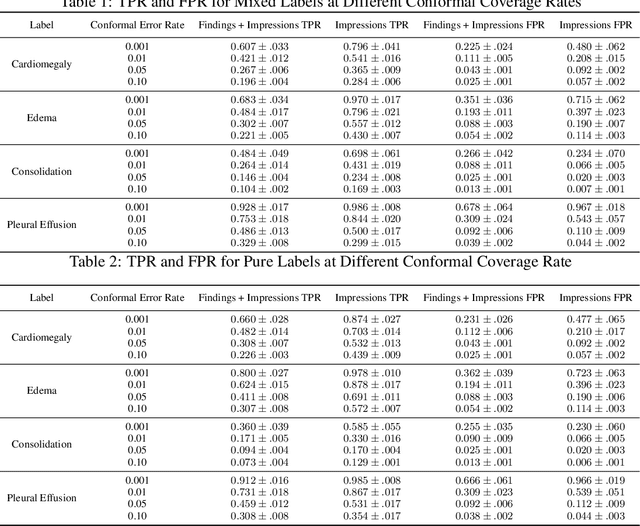
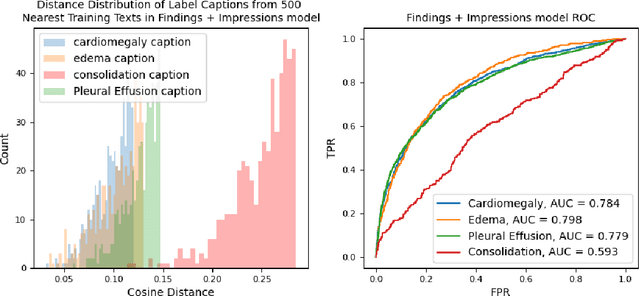
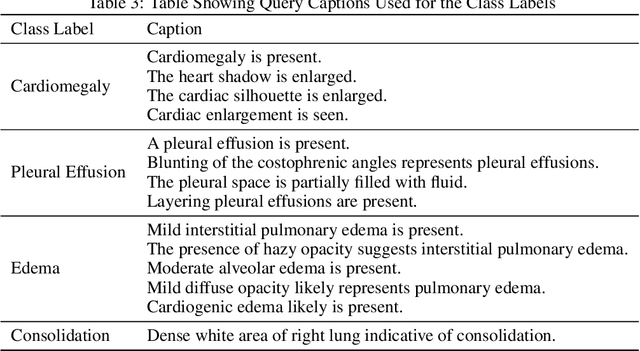
Abstract:Self-supervised models trained with a contrastive loss such as CLIP have shown to be very powerful in zero-shot classification settings. However, to be used as a zero-shot classifier these models require the user to provide new captions over a fixed set of labels at test time. In many settings, it is hard or impossible to know if a new query caption is compatible with the source captions used to train the model. We address these limitations by framing the zero-shot classification task as an outlier detection problem and develop a conformal prediction procedure to assess when a given test caption may be reliably used. On a real-world medical example, we show that our proposed conformal procedure improves the reliability of CLIP-style models in the zero-shot classification setting, and we provide an empirical analysis of the factors that may affect its performance.
Self-Supervision on Images and Text Reduces Reliance on Visual Shortcut Features
Jun 14, 2022

Abstract:Deep learning models trained in a fully supervised manner have been shown to rely on so-called "shortcut" features. Shortcut features are inputs that are associated with the outcome of interest in the training data, but are either no longer associated or not present in testing or deployment settings. Here we provide experiments that show recent self-supervised models trained on images and text provide more robust image representations and reduce the model's reliance on visual shortcut features on a realistic medical imaging example. Additionally, we find that these self-supervised models "forget" shortcut features more quickly than fully supervised ones when fine-tuned on labeled data. Though not a complete solution, our experiments provide compelling evidence that self-supervised models trained on images and text provide some resilience to visual shortcut features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge