Andrew L. Beam
DAG-aware Transformer for Causal Effect Estimation
Oct 13, 2024



Abstract:Causal inference is a critical task across fields such as healthcare, economics, and the social sciences. While recent advances in machine learning, especially those based on the deep-learning architectures, have shown potential in estimating causal effects, existing approaches often fall short in handling complex causal structures and lack adaptability across various causal scenarios. In this paper, we present a novel transformer-based method for causal inference that overcomes these challenges. The core innovation of our model lies in its integration of causal Directed Acyclic Graphs (DAGs) directly into the attention mechanism, enabling it to accurately model the underlying causal structure. This allows for flexible estimation of both average treatment effects (ATE) and conditional average treatment effects (CATE). Extensive experiments on both synthetic and real-world datasets demonstrate that our approach surpasses existing methods in estimating causal effects across a wide range of scenarios. The flexibility and robustness of our model make it a valuable tool for researchers and practitioners tackling complex causal inference problems.
TIER: Text-Image Entropy Regularization for CLIP-style models
Dec 13, 2022Abstract:In this paper, we study the effect of a novel regularization scheme on contrastive language-image pre-trained (CLIP) models. Our approach is based on the observation that, in many domains, text tokens should only describe a small number of image regions and, likewise, each image region should correspond to only a few text tokens. In CLIP-style models, this implies that text-token embeddings should have high similarity to only a small number of image-patch embeddings for a given image-text pair. We formalize this observation using a novel regularization scheme that penalizes the entropy of the text-token to image-patch similarity scores. We qualitatively and quantitatively demonstrate that the proposed regularization scheme shrinks the text-token and image-patch similarity scores towards zero, thus achieving the desired effect. We demonstrate the promise of our approach in an important medical context where this underlying hypothesis naturally arises. Using our proposed approach, we achieve state of the art (SOTA) zero-shot performance on all tasks from the CheXpert chest x-ray dataset, outperforming an unregularized version of the model and several recently published self-supervised models.
Deep Learning Methods for Proximal Inference via Maximum Moment Restriction
May 19, 2022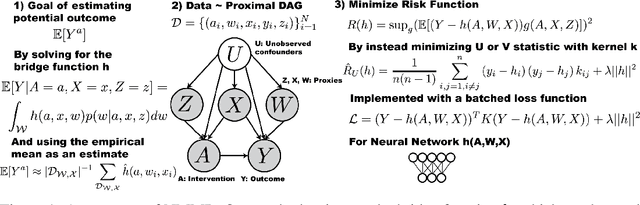
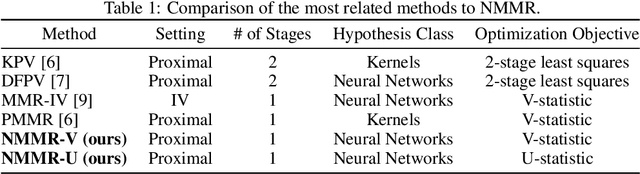
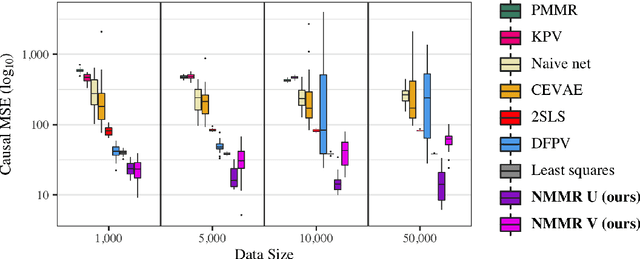
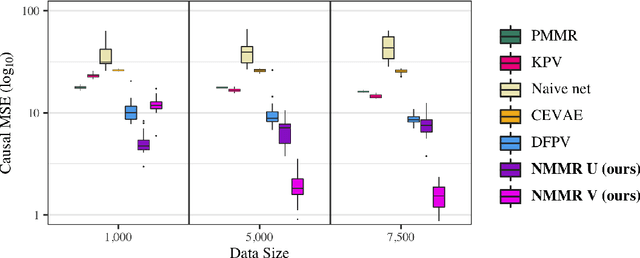
Abstract:The No Unmeasured Confounding Assumption is widely used to identify causal effects in observational studies. Recent work on proximal inference has provided alternative identification results that succeed even in the presence of unobserved confounders, provided that one has measured a sufficiently rich set of proxy variables, satisfying specific structural conditions. However, proximal inference requires solving an ill-posed integral equation. Previous approaches have used a variety of machine learning techniques to estimate a solution to this integral equation, commonly referred to as the bridge function. However, prior work has often been limited by relying on pre-specified kernel functions, which are not data adaptive and struggle to scale to large datasets. In this work, we introduce a flexible and scalable method based on a deep neural network to estimate causal effects in the presence of unmeasured confounding using proximal inference. Our method achieves state of the art performance on two well-established proximal inference benchmarks. Finally, we provide theoretical consistency guarantees for our method.
MedSelect: Selective Labeling for Medical Image Classification Combining Meta-Learning with Deep Reinforcement Learning
Mar 26, 2021


Abstract:We propose a selective learning method using meta-learning and deep reinforcement learning for medical image interpretation in the setting of limited labeling resources. Our method, MedSelect, consists of a trainable deep learning selector that uses image embeddings obtained from contrastive pretraining for determining which images to label, and a non-parametric selector that uses cosine similarity to classify unseen images. We demonstrate that MedSelect learns an effective selection strategy outperforming baseline selection strategies across seen and unseen medical conditions for chest X-ray interpretation. We also perform an analysis of the selections performed by MedSelect comparing the distribution of latent embeddings and clinical features, and find significant differences compared to the strongest performing baseline. We believe that our method may be broadly applicable across medical imaging settings where labels are expensive to acquire.
Evaluating Progress on Machine Learning for Longitudinal Electronic Healthcare Data
Oct 02, 2020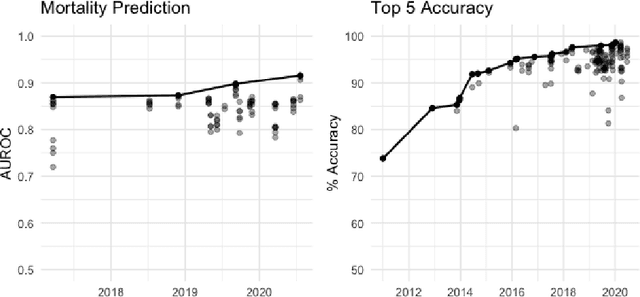
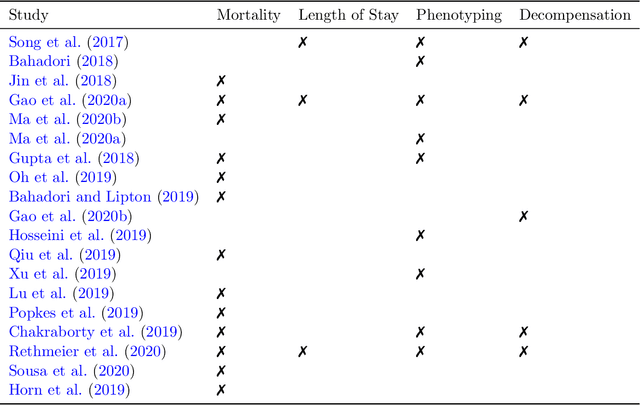
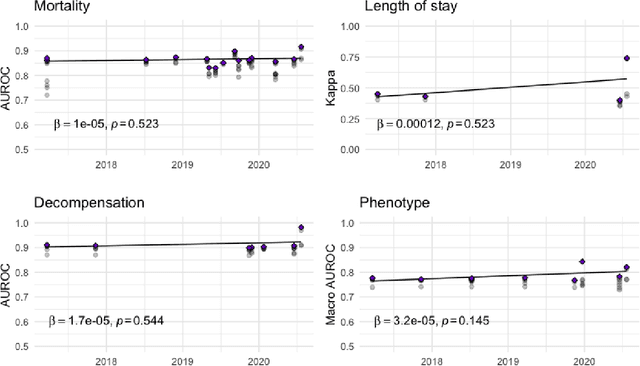
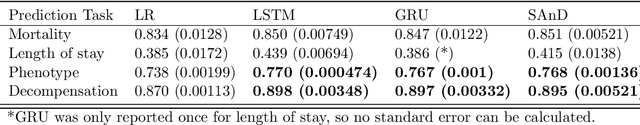
Abstract:The Large Scale Visual Recognition Challenge based on the well-known Imagenet dataset catalyzed an intense flurry of progress in computer vision. Benchmark tasks have propelled other sub-fields of machine learning forward at an equally impressive pace, but in healthcare it has primarily been image processing tasks, such as in dermatology and radiology, that have experienced similar benchmark-driven progress. In the present study, we performed a comprehensive review of benchmarks in medical machine learning for structured data, identifying one based on the Medical Information Mart for Intensive Care (MIMIC-III) that allows the first direct comparison of predictive performance and thus the evaluation of progress on four clinical prediction tasks: mortality, length of stay, phenotyping, and patient decompensation. We find that little meaningful progress has been made over a 3 year period on these tasks, despite significant community engagement. Through our meta-analysis, we find that the performance of deep recurrent models is only superior to logistic regression on certain tasks. We conclude with a synthesis of these results, possible explanations, and a list of desirable qualities for future benchmarks in medical machine learning.
Machine Learning for Health (ML4H) Workshop at NeurIPS 2018
Nov 24, 2018Abstract:This volume represents the accepted submissions from the Machine Learning for Health (ML4H) workshop at the conference on Neural Information Processing Systems (NeurIPS) 2018, held on December 8, 2018 in Montreal, Canada.
Learning Contextual Hierarchical Structure of Medical Concepts with Poincairé Embeddings to Clarify Phenotypes
Nov 03, 2018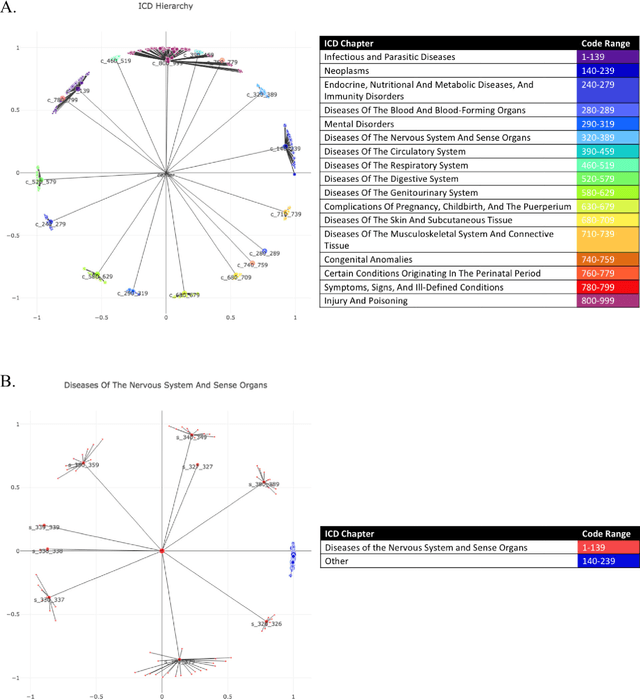
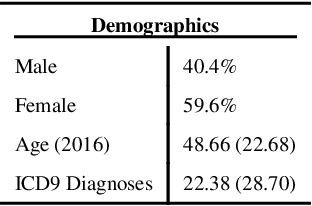
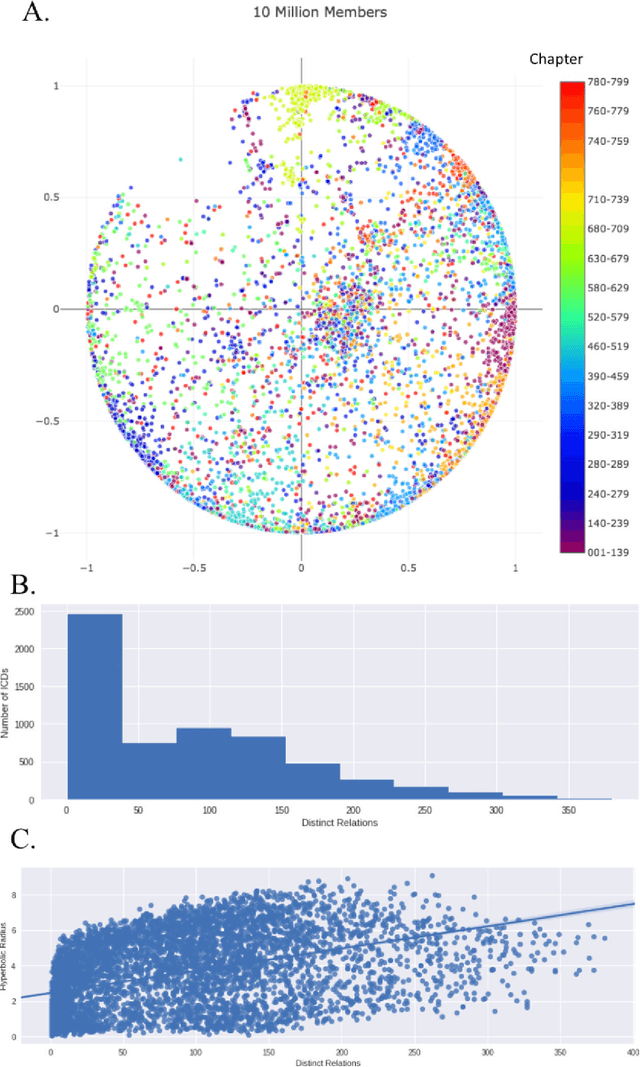
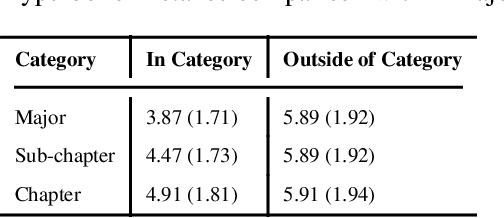
Abstract:Biomedical association studies are increasingly done using clinical concepts, and in particular diagnostic codes from clinical data repositories as phenotypes. Clinical concepts can be represented in a meaningful, vector space using word embedding models. These embeddings allow for comparison between clinical concepts or for straightforward input to machine learning models. Using traditional approaches, good representations require high dimensionality, making downstream tasks such as visualization more difficult. We applied Poincar\'e embeddings in a 2-dimensional hyperbolic space to a large-scale administrative claims database and show performance comparable to 100-dimensional embeddings in a euclidean space. We then examine disease relationships under different disease contexts to better understand potential phenotypes.
Opportunities in Machine Learning for Healthcare
Jun 05, 2018Abstract:Healthcare is a natural arena for the application of machine learning, especially as modern electronic health records (EHRs) provide increasingly large amounts of data to answer clinically meaningful questions. However, clinical data and practice present unique challenges that complicate the use of common methodologies. This article serves as a primer on addressing these challenges and highlights opportunities for members of the machine learning and data science communities to contribute to this growing domain.
Adversarial Attacks Against Medical Deep Learning Systems
May 21, 2018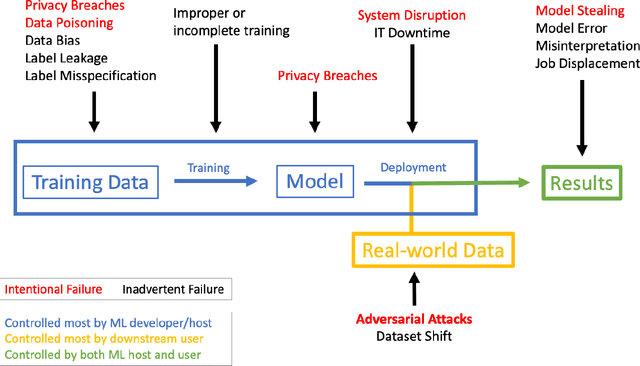
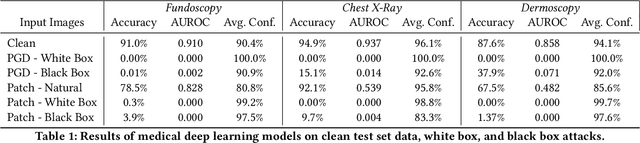
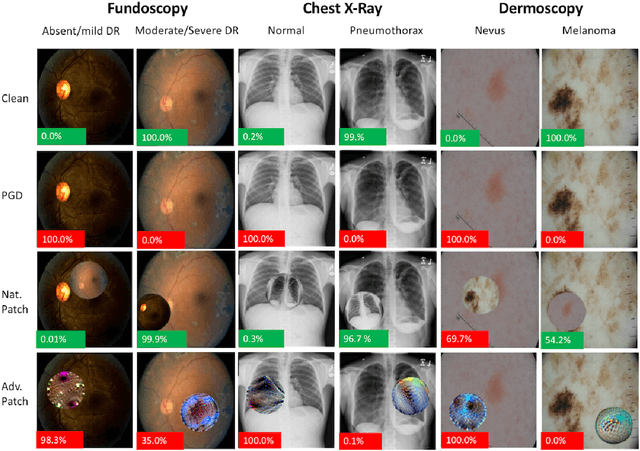
Abstract:The discovery of adversarial examples has raised concerns about the practical deployment of deep learning systems. In this paper, we argue that the field of medicine may be uniquely susceptible to adversarial attacks, both in terms of monetary incentives and technical vulnerability. To this end, we outline the healthcare economy and the incentives it creates for fraud, we extend adversarial attacks to three popular medical imaging tasks, and we provide concrete examples of how and why such attacks could be realistically carried out. For each of our representative medical deep learning classifiers, both white and black box attacks were highly successful. We urge caution in deploying deep learning systems in clinical settings, and encourage the machine learning community to further investigate the domain-specific characteristics of medical learning systems.
Clinical Concept Embeddings Learned from Massive Sources of Multimodal Medical Data
May 18, 2018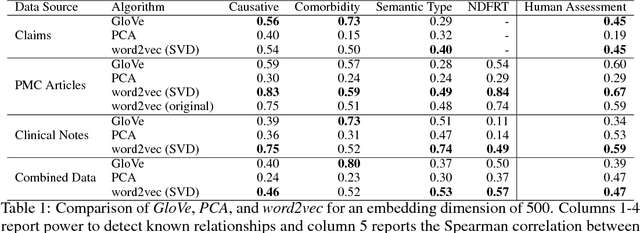
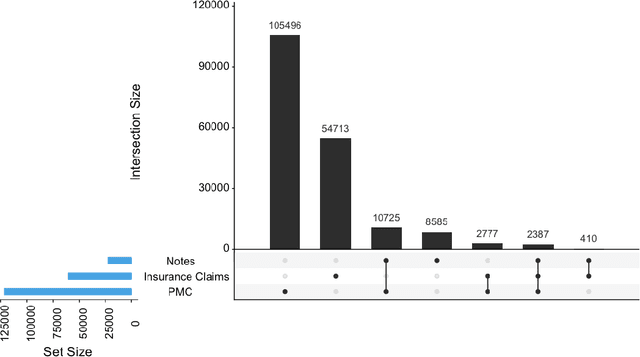
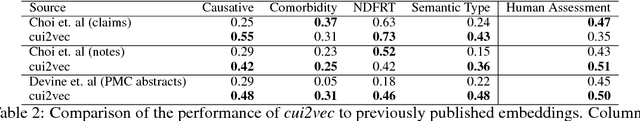
Abstract:Word embeddings are a popular approach to unsupervised learning of word relationships that are widely used in natural language processing. In this article, we present a new set of embeddings for medical concepts learned using an extremely large collection of multimodal medical data. Leaning on recent theoretical insights, we demonstrate how an insurance claims database of 60 million members, a collection of 20 million clinical notes, and 1.7 million full text biomedical journal articles can be combined to embed concepts into a common space, resulting in the largest ever set of embeddings for 108,477 medical concepts. To evaluate our approach, we present a new benchmark methodology based on statistical power specifically designed to test embeddings of medical concepts. Our approach, called cui2vec, attains state of the art performance relative to previous methods in most instances. Finally, we provide a downloadable set of pre-trained embeddings for other researchers to use, as well as an online tool for interactive exploration of the cui2vec embeddings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge