Isaac S. Kohane
One Patient, Many Contexts: Scaling Medical AI Through Contextual Intelligence
Jun 11, 2025

Abstract:Medical foundation models, including language models trained on clinical notes, vision-language models on medical images, and multimodal models on electronic health records, can summarize clinical notes, answer medical questions, and assist in decision-making. Adapting these models to new populations, specialties, or settings typically requires fine-tuning, careful prompting, or retrieval from knowledge bases. This can be impractical, and limits their ability to interpret unfamiliar inputs and adjust to clinical situations not represented during training. As a result, models are prone to contextual errors, where predictions appear reasonable but fail to account for critical patient-specific or contextual information. These errors stem from a fundamental limitation that current models struggle with: dynamically adjusting their behavior across evolving contexts of medical care. In this Perspective, we outline a vision for context-switching in medical AI: models that dynamically adapt their reasoning without retraining to new specialties, populations, workflows, and clinical roles. We envision context-switching AI to diagnose, manage, and treat a wide range of diseases across specialties and regions, and expand access to medical care.
Causal machine learning for predicting treatment outcomes
Oct 11, 2024Abstract:Causal machine learning (ML) offers flexible, data-driven methods for predicting treatment outcomes including efficacy and toxicity, thereby supporting the assessment and safety of drugs. A key benefit of causal ML is that it allows for estimating individualized treatment effects, so that clinical decision-making can be personalized to individual patient profiles. Causal ML can be used in combination with both clinical trial data and real-world data, such as clinical registries and electronic health records, but caution is needed to avoid biased or incorrect predictions. In this Perspective, we discuss the benefits of causal ML (relative to traditional statistical or ML approaches) and outline the key components and steps. Finally, we provide recommendations for the reliable use of causal ML and effective translation into the clinic.
* Accepted version; not Version of Record
Identifying Heterogeneous Treatment Effects in Multiple Outcomes using Joint Confidence Intervals
Dec 02, 2022


Abstract:Heterogeneous treatment effects (HTEs) are commonly identified during randomized controlled trials (RCTs). Identifying subgroups of patients with similar treatment effects is of high interest in clinical research to advance precision medicine. Often, multiple clinical outcomes are measured during an RCT, each having a potentially heterogeneous effect. Recently there has been high interest in identifying subgroups from HTEs, however, there has been less focus on developing tools in settings where there are multiple outcomes. In this work, we propose a framework for partitioning the covariate space to identify subgroups across multiple outcomes based on the joint CIs. We test our algorithm on synthetic and semi-synthetic data where there are two outcomes, and demonstrate that our algorithm is able to capture the HTE in both outcomes simultaneously.
* Accepted to ML4H 2022. Available at https://proceedings.mlr.press/v193/argaw22a.html
Approaching Small Molecule Prioritization as a Cross-Modal Information Retrieval Task through Coordinated Representation Learning
Nov 22, 2019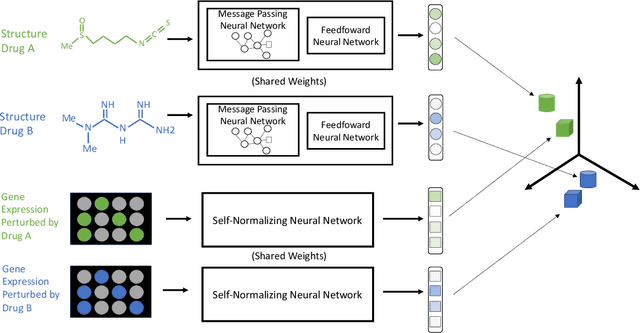
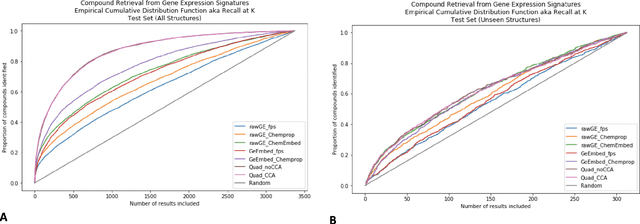
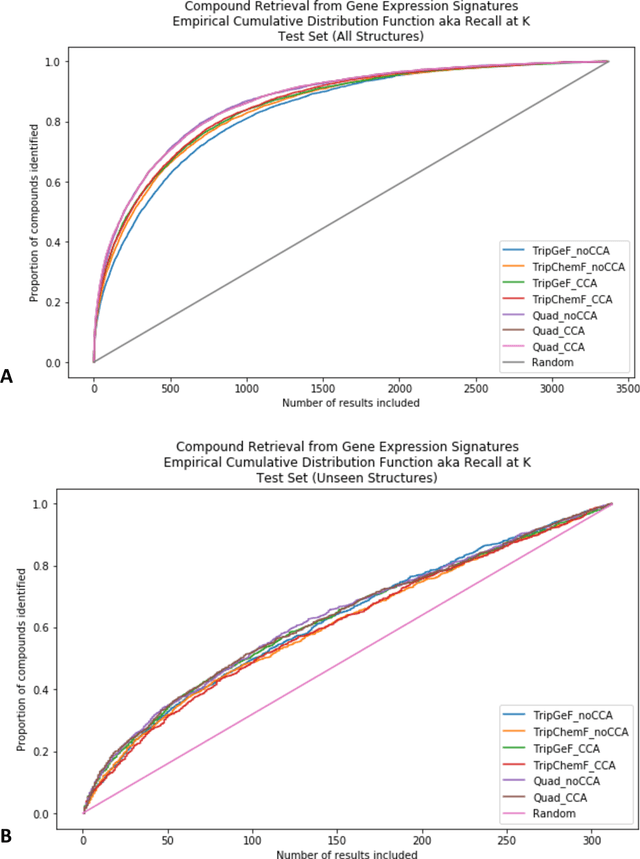
Abstract:Modeling the relationship between chemical structure and molecular activity is a key task in drug development and precision medicine. In this paper, we utilize a novel deep learning architecture to jointly train coordinated embeddings of chemical structures and transcriptional signatures. We do so by training neural networks in a coordinated manner such that learned chemical representations correlate most highly with the encodings of the transcriptional patterns they induce. We then test this approach by using held-out gene expression signatures as queries into embedding space to recover their corresponding compounds. We evaluate these embeddings' utility for small molecule prioritization on this new benchmark task. Our method outperforms a series of baselines, successfully generalizing to unseen transcriptional experiments, but still struggles to generalize to entirely unseen chemical structures.
Towards generative adversarial networks as a new paradigm for radiology education
Dec 04, 2018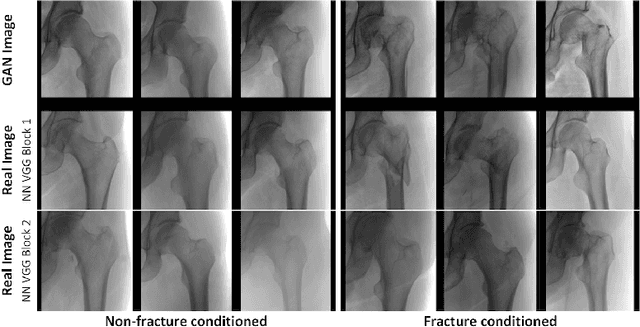
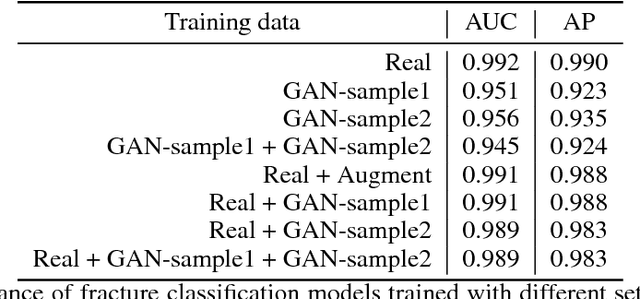
Abstract:Medical students and radiology trainees typically view thousands of images in order to "train their eye" to detect the subtle visual patterns necessary for diagnosis. Nevertheless, infrastructural and legal constraints often make it difficult to access and quickly query an abundance of images with a user-specified feature set. In this paper, we use a conditional generative adversarial network (GAN) to synthesize $1024\times1024$ pixel pelvic radiographs that can be queried with conditioning on fracture status. We demonstrate that the conditional GAN learns features that distinguish fractures from non-fractures by training a convolutional neural network exclusively on images sampled from the GAN and achieving an AUC of $>0.95$ on a held-out set of real images. We conduct additional analysis of the images sampled from the GAN and describe ongoing work to validate educational efficacy.
Learning Contextual Hierarchical Structure of Medical Concepts with Poincairé Embeddings to Clarify Phenotypes
Nov 03, 2018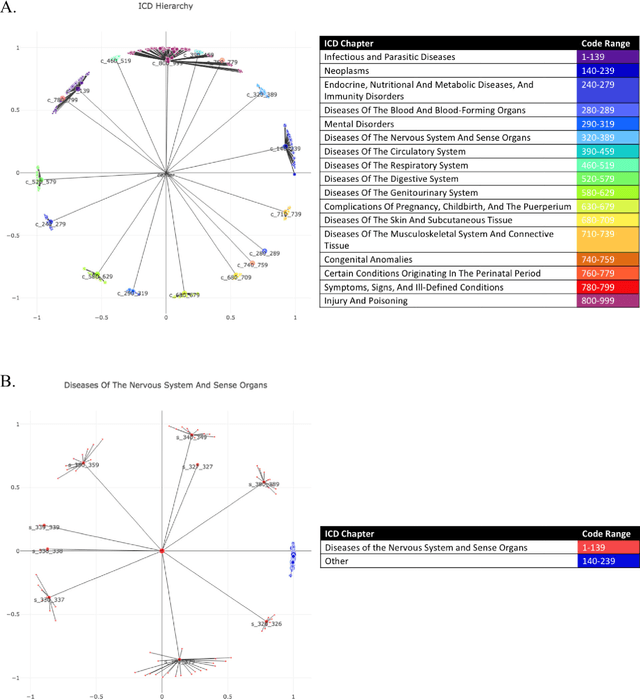
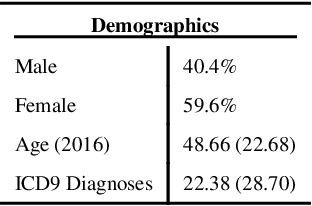
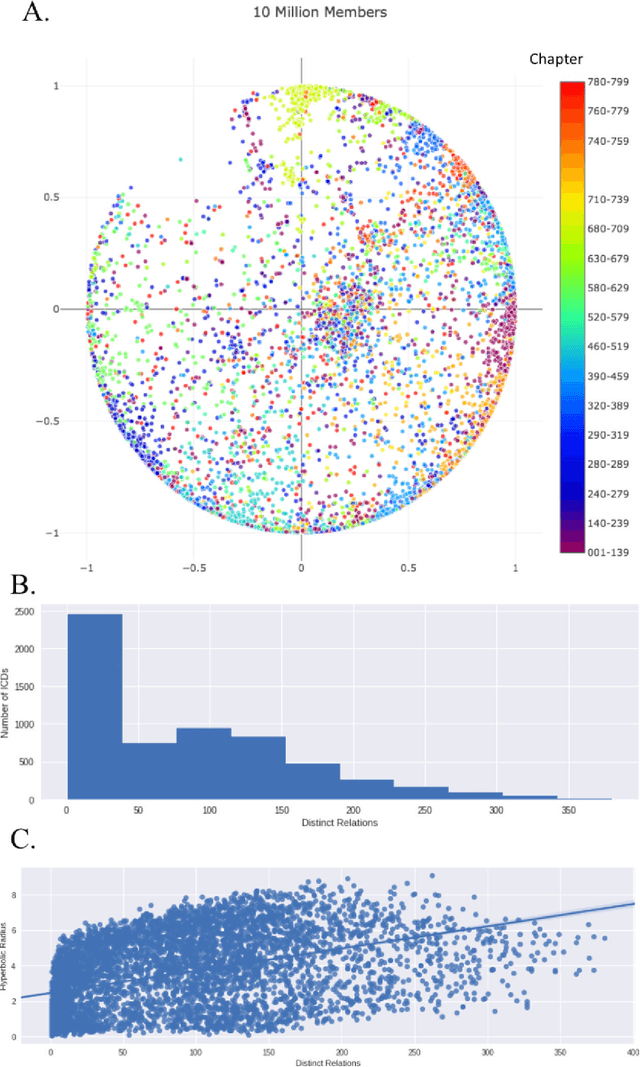
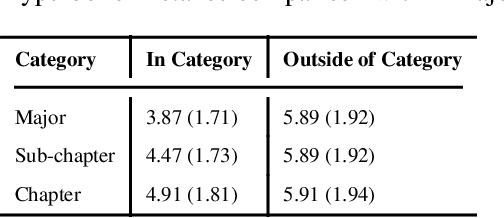
Abstract:Biomedical association studies are increasingly done using clinical concepts, and in particular diagnostic codes from clinical data repositories as phenotypes. Clinical concepts can be represented in a meaningful, vector space using word embedding models. These embeddings allow for comparison between clinical concepts or for straightforward input to machine learning models. Using traditional approaches, good representations require high dimensionality, making downstream tasks such as visualization more difficult. We applied Poincar\'e embeddings in a 2-dimensional hyperbolic space to a large-scale administrative claims database and show performance comparable to 100-dimensional embeddings in a euclidean space. We then examine disease relationships under different disease contexts to better understand potential phenotypes.
Adversarial Attacks Against Medical Deep Learning Systems
May 21, 2018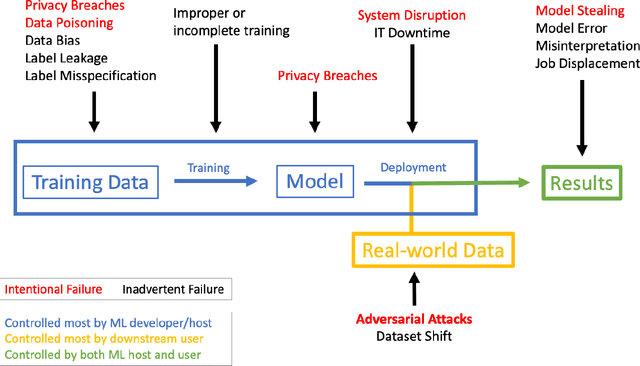
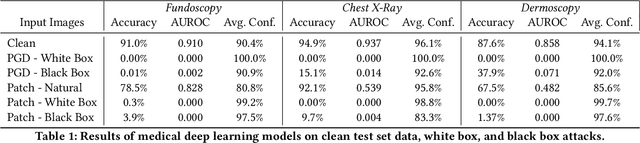
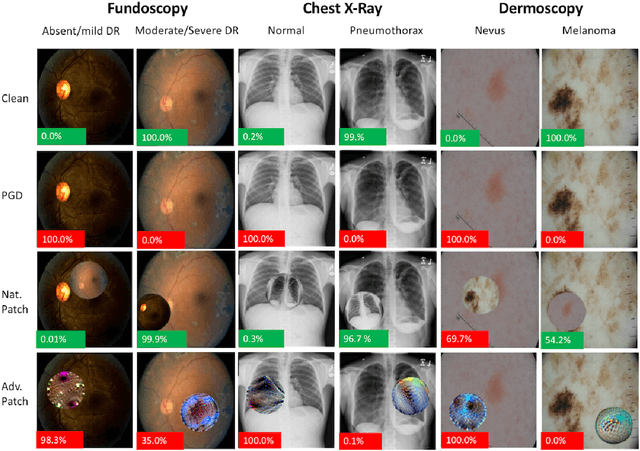
Abstract:The discovery of adversarial examples has raised concerns about the practical deployment of deep learning systems. In this paper, we argue that the field of medicine may be uniquely susceptible to adversarial attacks, both in terms of monetary incentives and technical vulnerability. To this end, we outline the healthcare economy and the incentives it creates for fraud, we extend adversarial attacks to three popular medical imaging tasks, and we provide concrete examples of how and why such attacks could be realistically carried out. For each of our representative medical deep learning classifiers, both white and black box attacks were highly successful. We urge caution in deploying deep learning systems in clinical settings, and encourage the machine learning community to further investigate the domain-specific characteristics of medical learning systems.
Clinical Concept Embeddings Learned from Massive Sources of Multimodal Medical Data
May 18, 2018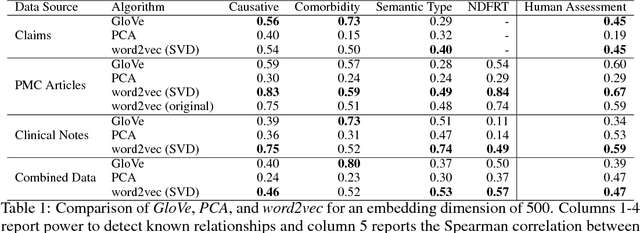
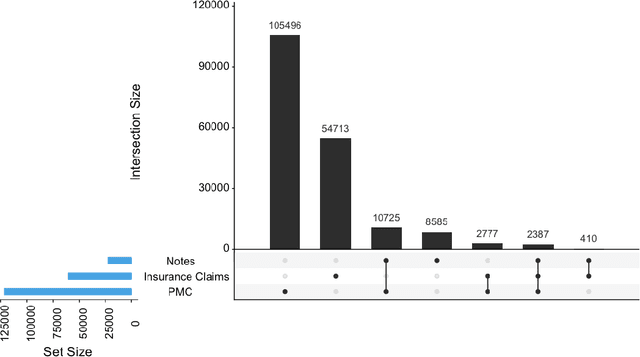
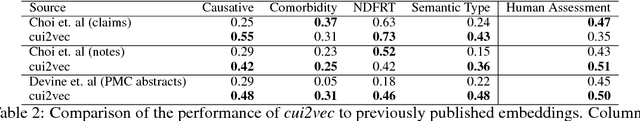
Abstract:Word embeddings are a popular approach to unsupervised learning of word relationships that are widely used in natural language processing. In this article, we present a new set of embeddings for medical concepts learned using an extremely large collection of multimodal medical data. Leaning on recent theoretical insights, we demonstrate how an insurance claims database of 60 million members, a collection of 20 million clinical notes, and 1.7 million full text biomedical journal articles can be combined to embed concepts into a common space, resulting in the largest ever set of embeddings for 108,477 medical concepts. To evaluate our approach, we present a new benchmark methodology based on statistical power specifically designed to test embeddings of medical concepts. Our approach, called cui2vec, attains state of the art performance relative to previous methods in most instances. Finally, we provide a downloadable set of pre-trained embeddings for other researchers to use, as well as an online tool for interactive exploration of the cui2vec embeddings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge