Jonas Schweisthal
LLM-Driven Treatment Effect Estimation Under Inference Time Text Confounding
Jul 03, 2025Abstract:Estimating treatment effects is crucial for personalized decision-making in medicine, but this task faces unique challenges in clinical practice. At training time, models for estimating treatment effects are typically trained on well-structured medical datasets that contain detailed patient information. However, at inference time, predictions are often made using textual descriptions (e.g., descriptions with self-reported symptoms), which are incomplete representations of the original patient information. In this work, we make three contributions. (1) We show that the discrepancy between the data available during training time and inference time can lead to biased estimates of treatment effects. We formalize this issue as an inference time text confounding problem, where confounders are fully observed during training time but only partially available through text at inference time. (2) To address this problem, we propose a novel framework for estimating treatment effects that explicitly accounts for inference time text confounding. Our framework leverages large language models together with a custom doubly robust learner to mitigate biases caused by the inference time text confounding. (3) Through a series of experiments, we demonstrate the effectiveness of our framework in real-world applications.
Treatment Effect Estimation for Optimal Decision-Making
May 19, 2025Abstract:Decision-making across various fields, such as medicine, heavily relies on conditional average treatment effects (CATEs). Practitioners commonly make decisions by checking whether the estimated CATE is positive, even though the decision-making performance of modern CATE estimators is poorly understood from a theoretical perspective. In this paper, we study optimal decision-making based on two-stage CATE estimators (e.g., DR-learner), which are considered state-of-the-art and widely used in practice. We prove that, while such estimators may be optimal for estimating CATE, they can be suboptimal when used for decision-making. Intuitively, this occurs because such estimators prioritize CATE accuracy in regions far away from the decision boundary, which is ultimately irrelevant to decision-making. As a remedy, we propose a novel two-stage learning objective that retargets the CATE to balance CATE estimation error and decision performance. We then propose a neural method that optimizes an adaptively-smoothed approximation of our learning objective. Finally, we confirm the effectiveness of our method both empirically and theoretically. In sum, our work is the first to show how two-stage CATE estimators can be adapted for optimal decision-making.
Orthogonal Representation Learning for Estimating Causal Quantities
Feb 06, 2025Abstract:Representation learning is widely used for estimating causal quantities (e.g., the conditional average treatment effect) from observational data. While existing representation learning methods have the benefit of allowing for end-to-end learning, they do not have favorable theoretical properties of Neyman-orthogonal learners, such as double robustness and quasi-oracle efficiency. Also, such representation learning methods often employ additional constraints, like balancing, which may even lead to inconsistent estimation. In this paper, we propose a novel class of Neyman-orthogonal learners for causal quantities defined at the representation level, which we call OR-learners. Our OR-learners have several practical advantages: they allow for consistent estimation of causal quantities based on any learned representation, while offering favorable theoretical properties including double robustness and quasi-oracle efficiency. In multiple experiments, we show that, under certain regularity conditions, our OR-learners improve existing representation learning methods and achieve state-of-the-art performance. To the best of our knowledge, our OR-learners are the first work to offer a unified framework of representation learning methods and Neyman-orthogonal learners for causal quantities estimation.
Constructing Confidence Intervals for Average Treatment Effects from Multiple Datasets
Dec 16, 2024



Abstract:Constructing confidence intervals (CIs) for the average treatment effect (ATE) from patient records is crucial to assess the effectiveness and safety of drugs. However, patient records typically come from different hospitals, thus raising the question of how multiple observational datasets can be effectively combined for this purpose. In our paper, we propose a new method that estimates the ATE from multiple observational datasets and provides valid CIs. Our method makes little assumptions about the observational datasets and is thus widely applicable in medical practice. The key idea of our method is that we leverage prediction-powered inferences and thereby essentially `shrink' the CIs so that we offer more precise uncertainty quantification as compared to na\"ive approaches. We further prove the unbiasedness of our method and the validity of our CIs. We confirm our theoretical results through various numerical experiments. Finally, we provide an extension of our method for constructing CIs from combinations of experimental and observational datasets.
DiffPO: A causal diffusion model for learning distributions of potential outcomes
Oct 11, 2024



Abstract:Predicting potential outcomes of interventions from observational data is crucial for decision-making in medicine, but the task is challenging due to the fundamental problem of causal inference. Existing methods are largely limited to point estimates of potential outcomes with no uncertain quantification; thus, the full information about the distributions of potential outcomes is typically ignored. In this paper, we propose a novel causal diffusion model called DiffPO, which is carefully designed for reliable inferences in medicine by learning the distribution of potential outcomes. In our DiffPO, we leverage a tailored conditional denoising diffusion model to learn complex distributions, where we address the selection bias through a novel orthogonal diffusion loss. Another strength of our DiffPO method is that it is highly flexible (e.g., it can also be used to estimate different causal quantities such as CATE). Across a wide range of experiments, we show that our method achieves state-of-the-art performance.
Learning Representations of Instruments for Partial Identification of Treatment Effects
Oct 11, 2024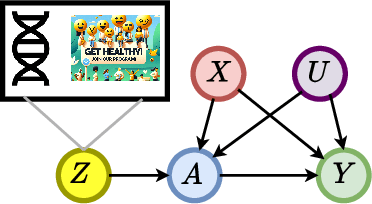
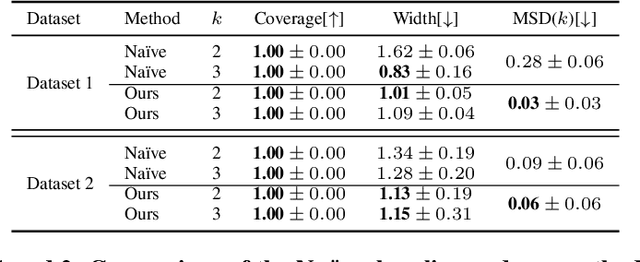
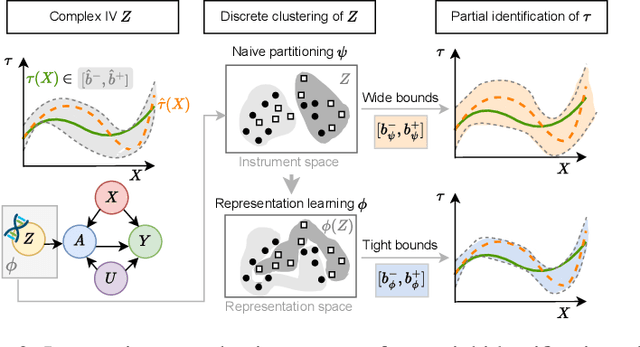
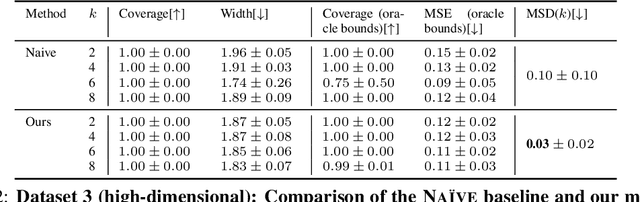
Abstract:Reliable estimation of treatment effects from observational data is important in many disciplines such as medicine. However, estimation is challenging when unconfoundedness as a standard assumption in the causal inference literature is violated. In this work, we leverage arbitrary (potentially high-dimensional) instruments to estimate bounds on the conditional average treatment effect (CATE). Our contributions are three-fold: (1) We propose a novel approach for partial identification through a mapping of instruments to a discrete representation space so that we yield valid bounds on the CATE. This is crucial for reliable decision-making in real-world applications. (2) We derive a two-step procedure that learns tight bounds using a tailored neural partitioning of the latent instrument space. As a result, we avoid instability issues due to numerical approximations or adversarial training. Furthermore, our procedure aims to reduce the estimation variance in finite-sample settings to yield more reliable estimates. (3) We show theoretically that our procedure obtains valid bounds while reducing estimation variance. We further perform extensive experiments to demonstrate the effectiveness across various settings. Overall, our procedure offers a novel path for practitioners to make use of potentially high-dimensional instruments (e.g., as in Mendelian randomization).
Causal machine learning for predicting treatment outcomes
Oct 11, 2024Abstract:Causal machine learning (ML) offers flexible, data-driven methods for predicting treatment outcomes including efficacy and toxicity, thereby supporting the assessment and safety of drugs. A key benefit of causal ML is that it allows for estimating individualized treatment effects, so that clinical decision-making can be personalized to individual patient profiles. Causal ML can be used in combination with both clinical trial data and real-world data, such as clinical registries and electronic health records, but caution is needed to avoid biased or incorrect predictions. In this Perspective, we discuss the benefits of causal ML (relative to traditional statistical or ML approaches) and outline the key components and steps. Finally, we provide recommendations for the reliable use of causal ML and effective translation into the clinic.
* Accepted version; not Version of Record
Conformal Prediction for Causal Effects of Continuous Treatments
Jul 03, 2024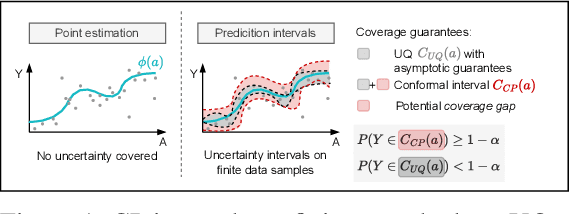

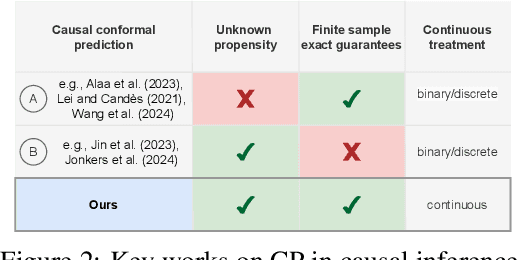

Abstract:Uncertainty quantification of causal effects is crucial for safety-critical applications such as personalized medicine. A powerful approach for this is conformal prediction, which has several practical benefits due to model-agnostic finite-sample guarantees. Yet, existing methods for conformal prediction of causal effects are limited to binary/discrete treatments and make highly restrictive assumptions such as known propensity scores. In this work, we provide a novel conformal prediction method for potential outcomes of continuous treatments. We account for the additional uncertainty introduced through propensity estimation so that our conformal prediction intervals are valid even if the propensity score is unknown. Our contributions are three-fold: (1) We derive finite-sample prediction intervals for potential outcomes of continuous treatments. (2) We provide an algorithm for calculating the derived intervals. (3) We demonstrate the effectiveness of the conformal prediction intervals in experiments on synthetic and real-world datasets. To the best of our knowledge, we are the first to propose conformal prediction for continuous treatments when the propensity score is unknown and must be estimated from data.
Meta-Learners for Partially-Identified Treatment Effects Across Multiple Environments
Jun 04, 2024



Abstract:Estimating the conditional average treatment effect (CATE) from observational data is relevant for many applications such as personalized medicine. Here, we focus on the widespread setting where the observational data come from multiple environments, such as different hospitals, physicians, or countries. Furthermore, we allow for violations of standard causal assumptions, namely, overlap within the environments and unconfoundedness. To this end, we move away from point identification and focus on partial identification. Specifically, we show that current assumptions from the literature on multiple environments allow us to interpret the environment as an instrumental variable (IV). This allows us to adapt bounds from the IV literature for partial identification of CATE by leveraging treatment assignment mechanisms across environments. Then, we propose different model-agnostic learners (so-called meta-learners) to estimate the bounds that can be used in combination with arbitrary machine learning models. We further demonstrate the effectiveness of our meta-learners across various experiments using both simulated and real-world data. Finally, we discuss the applicability of our meta-learners to partial identification in instrumental variable settings, such as randomized controlled trials with non-compliance.
Reliable Off-Policy Learning for Dosage Combinations
May 31, 2023Abstract:Decision-making in personalized medicine such as cancer therapy or critical care must often make choices for dosage combinations, i.e., multiple continuous treatments. Existing work for this task has modeled the effect of multiple treatments independently, while estimating the joint effect has received little attention but comes with non-trivial challenges. In this paper, we propose a novel method for reliable off-policy learning for dosage combinations. Our method proceeds along three steps: (1) We develop a tailored neural network that estimates the individualized dose-response function while accounting for the joint effect of multiple dependent dosages. (2) We estimate the generalized propensity score using conditional normalizing flows in order to detect regions with limited overlap in the shared covariate-treatment space. (3) We present a gradient-based learning algorithm to find the optimal, individualized dosage combinations. Here, we ensure reliable estimation of the policy value by avoiding regions with limited overlap. We finally perform an extensive evaluation of our method to show its effectiveness. To the best of our knowledge, ours is the first work to provide a method for reliable off-policy learning for optimal dosage combinations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge