Scott L. Lipnick
Approaching Small Molecule Prioritization as a Cross-Modal Information Retrieval Task through Coordinated Representation Learning
Nov 22, 2019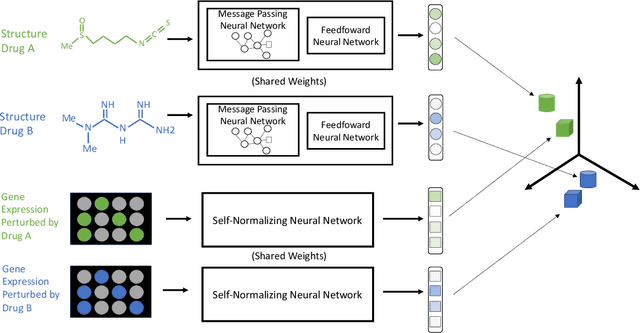
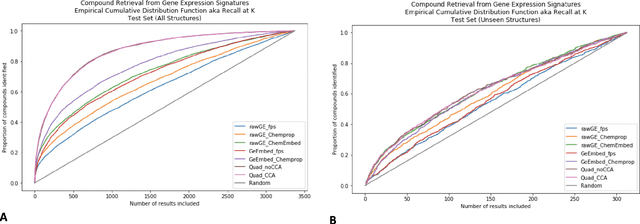
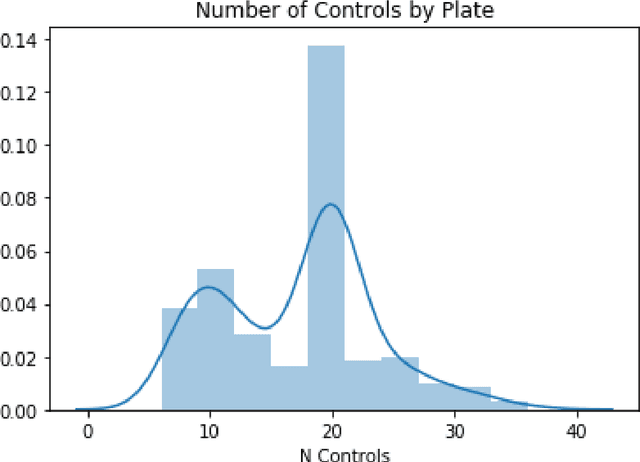
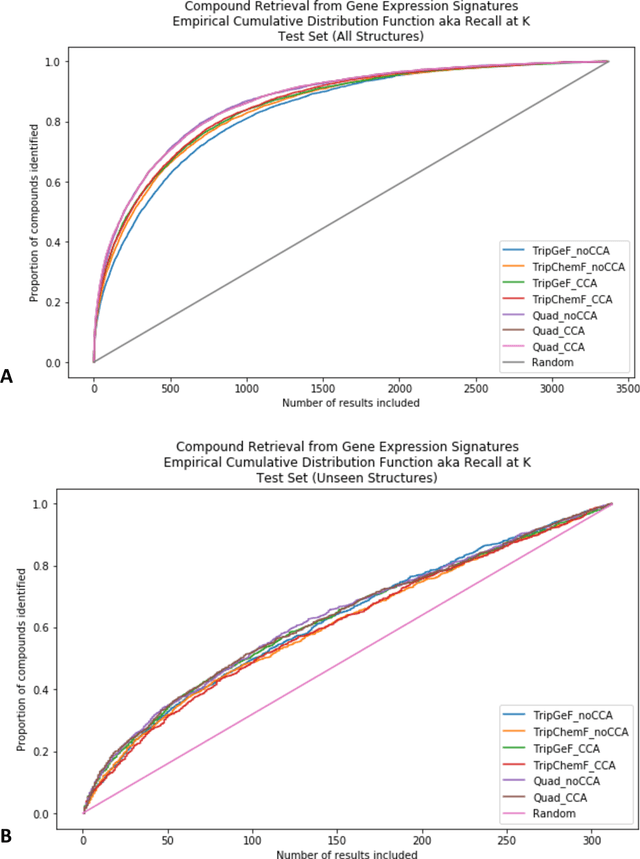
Abstract:Modeling the relationship between chemical structure and molecular activity is a key task in drug development and precision medicine. In this paper, we utilize a novel deep learning architecture to jointly train coordinated embeddings of chemical structures and transcriptional signatures. We do so by training neural networks in a coordinated manner such that learned chemical representations correlate most highly with the encodings of the transcriptional patterns they induce. We then test this approach by using held-out gene expression signatures as queries into embedding space to recover their corresponding compounds. We evaluate these embeddings' utility for small molecule prioritization on this new benchmark task. Our method outperforms a series of baselines, successfully generalizing to unseen transcriptional experiments, but still struggles to generalize to entirely unseen chemical structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge