Samuel G. Finlayson
Subgraph Neural Networks
Jun 19, 2020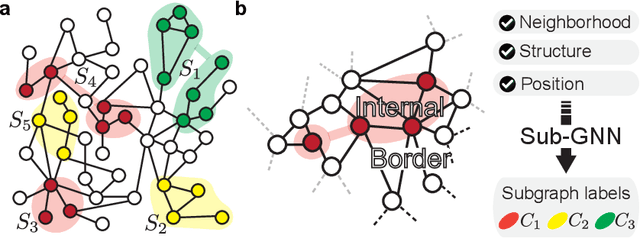
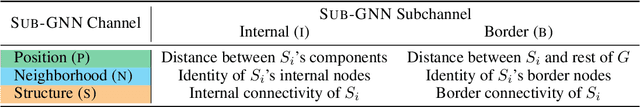
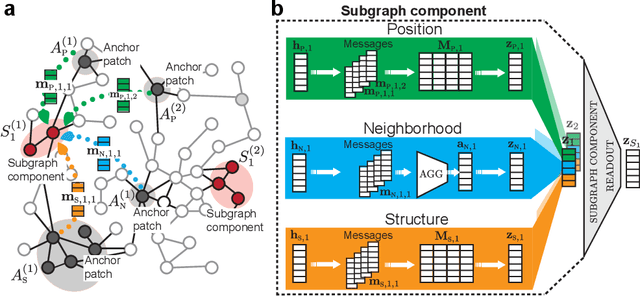
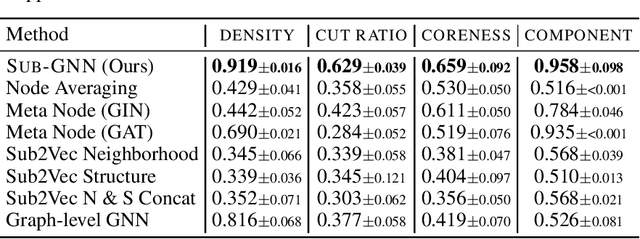
Abstract:Deep learning methods for graphs achieve remarkable performance on many node-level and graph-level prediction tasks. However, despite the proliferation of the methods and their success, prevailing Graph Neural Networks (GNNs) neglect subgraphs, rendering subgraph prediction tasks challenging to tackle in many impactful applications. Further, subgraph prediction tasks present several unique challenges, because subgraphs can have non-trivial internal topology, but also carry a notion of position and external connectivity information relative to the underlying graph in which they exist. Here, we introduce SUB-GNN, a subgraph neural network to learn disentangled subgraph representations. In particular, we propose a novel subgraph routing mechanism that propagates neural messages between the subgraph's components and randomly sampled anchor patches from the underlying graph, yielding highly accurate subgraph representations. SUB-GNN specifies three channels, each designed to capture a distinct aspect of subgraph structure, and we provide empirical evidence that the channels encode their intended properties. We design a series of new synthetic and real-world subgraph datasets. Empirical results for subgraph classification on eight datasets show that SUB-GNN achieves considerable performance gains, outperforming strong baseline methods, including node-level and graph-level GNNs, by 12.4% over the strongest baseline. SUB-GNN performs exceptionally well on challenging biomedical datasets when subgraphs have complex topology and even comprise multiple disconnected components.
Approaching Small Molecule Prioritization as a Cross-Modal Information Retrieval Task through Coordinated Representation Learning
Nov 22, 2019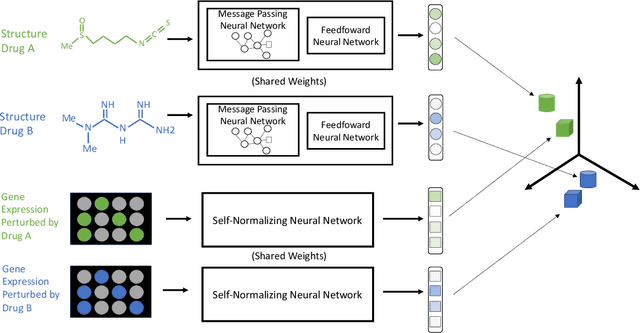
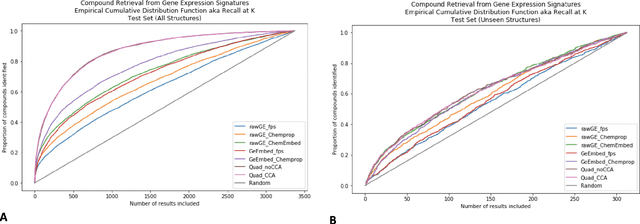
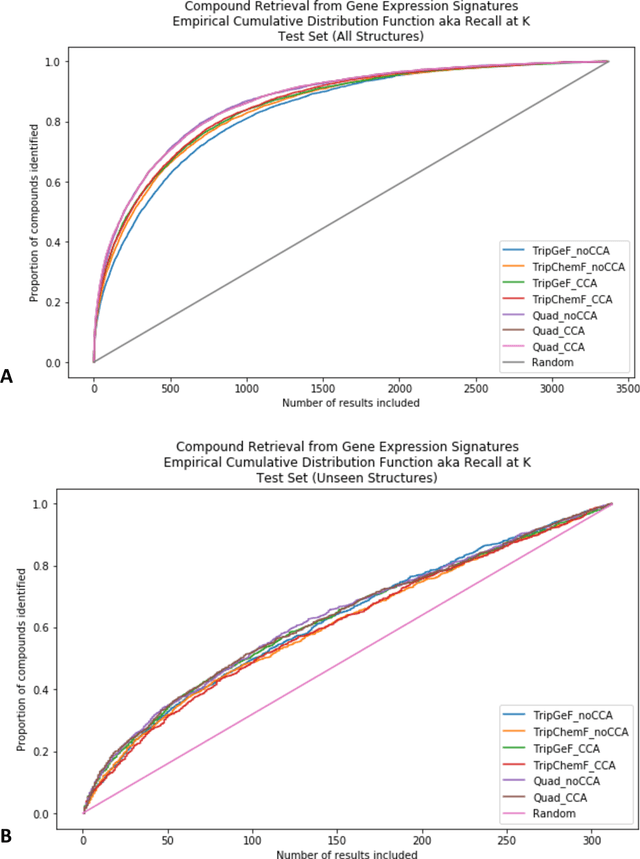
Abstract:Modeling the relationship between chemical structure and molecular activity is a key task in drug development and precision medicine. In this paper, we utilize a novel deep learning architecture to jointly train coordinated embeddings of chemical structures and transcriptional signatures. We do so by training neural networks in a coordinated manner such that learned chemical representations correlate most highly with the encodings of the transcriptional patterns they induce. We then test this approach by using held-out gene expression signatures as queries into embedding space to recover their corresponding compounds. We evaluate these embeddings' utility for small molecule prioritization on this new benchmark task. Our method outperforms a series of baselines, successfully generalizing to unseen transcriptional experiments, but still struggles to generalize to entirely unseen chemical structures.
Towards generative adversarial networks as a new paradigm for radiology education
Dec 04, 2018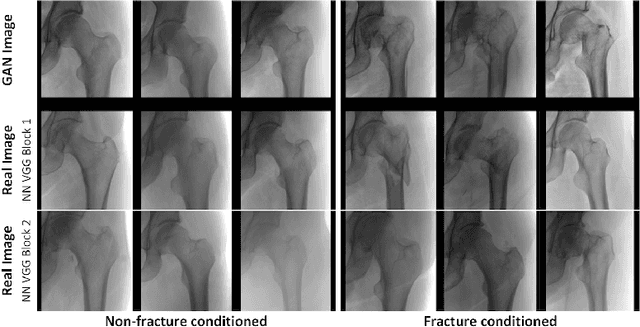
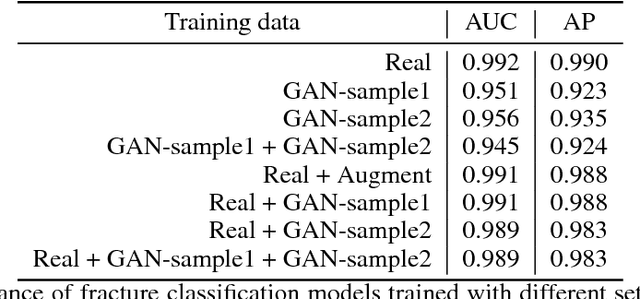
Abstract:Medical students and radiology trainees typically view thousands of images in order to "train their eye" to detect the subtle visual patterns necessary for diagnosis. Nevertheless, infrastructural and legal constraints often make it difficult to access and quickly query an abundance of images with a user-specified feature set. In this paper, we use a conditional generative adversarial network (GAN) to synthesize $1024\times1024$ pixel pelvic radiographs that can be queried with conditioning on fracture status. We demonstrate that the conditional GAN learns features that distinguish fractures from non-fractures by training a convolutional neural network exclusively on images sampled from the GAN and achieving an AUC of $>0.95$ on a held-out set of real images. We conduct additional analysis of the images sampled from the GAN and describe ongoing work to validate educational efficacy.
Privacy-Preserving Distributed Deep Learning for Clinical Data
Dec 04, 2018Abstract:Deep learning with medical data often requires larger samples sizes than are available at single providers. While data sharing among institutions is desirable to train more accurate and sophisticated models, it can lead to severe privacy concerns due the sensitive nature of the data. This problem has motivated a number of studies on distributed training of neural networks that do not require direct sharing of the training data. However, simple distributed training does not offer provable privacy guarantees to satisfy technical safe standards and may reveal information about the underlying patients. We present a method to train neural networks for clinical data in a distributed fashion under differential privacy. We demonstrate these methods on two datasets that include information from multiple independent sites, the eICU collaborative Research Database and The Cancer Genome Atlas.
Adversarial Attacks Against Medical Deep Learning Systems
May 21, 2018
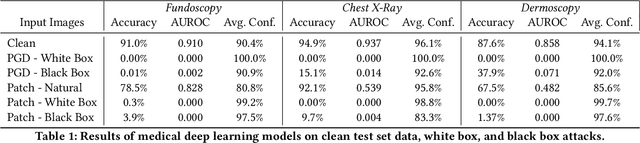
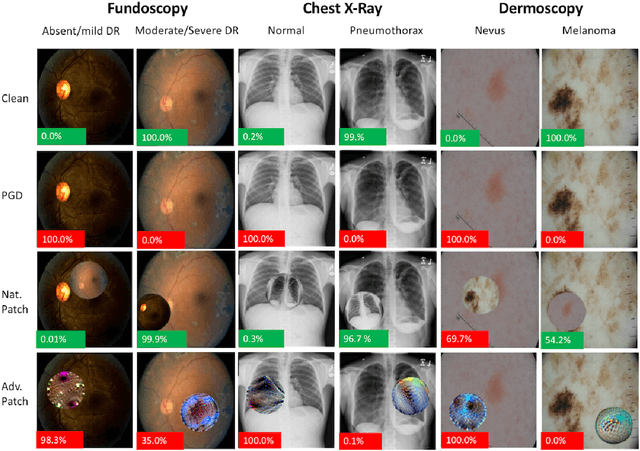
Abstract:The discovery of adversarial examples has raised concerns about the practical deployment of deep learning systems. In this paper, we argue that the field of medicine may be uniquely susceptible to adversarial attacks, both in terms of monetary incentives and technical vulnerability. To this end, we outline the healthcare economy and the incentives it creates for fraud, we extend adversarial attacks to three popular medical imaging tasks, and we provide concrete examples of how and why such attacks could be realistically carried out. For each of our representative medical deep learning classifiers, both white and black box attacks were highly successful. We urge caution in deploying deep learning systems in clinical settings, and encourage the machine learning community to further investigate the domain-specific characteristics of medical learning systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge