Emily Alsentzer
Massachusetts Institute of Technology, Harvard Medical School
AI-generated data contamination erodes pathological variability and diagnostic reliability
Jan 21, 2026Abstract:Generative artificial intelligence (AI) is rapidly populating medical records with synthetic content, creating a feedback loop where future models are increasingly at risk of training on uncurated AI-generated data. However, the clinical consequences of this AI-generated data contamination remain unexplored. Here, we show that in the absence of mandatory human verification, this self-referential cycle drives a rapid erosion of pathological variability and diagnostic reliability. By analysing more than 800,000 synthetic data points across clinical text generation, vision-language reporting, and medical image synthesis, we find that models progressively converge toward generic phenotypes regardless of the model architecture. Specifically, rare but critical findings, including pneumothorax and effusions, vanish from the synthetic content generated by AI models, while demographic representations skew heavily toward middle-aged male phenotypes. Crucially, this degradation is masked by false diagnostic confidence; models continue to issue reassuring reports while failing to detect life-threatening pathology, with false reassurance rates tripling to 40%. Blinded physician evaluation confirms that this decoupling of confidence and accuracy renders AI-generated documentation clinically useless after just two generations. We systematically evaluate three mitigation strategies, finding that while synthetic volume scaling fails to prevent collapse, mixing real data with quality-aware filtering effectively preserves diversity. Ultimately, our results suggest that without policy-mandated human oversight, the deployment of generative AI threatens to degrade the very healthcare data ecosystems it relies upon.
Large Language Models for Large-Scale, Rigorous Qualitative Analysis in Applied Health Services Research
Jan 20, 2026Abstract:Large language models (LLMs) show promise for improving the efficiency of qualitative analysis in large, multi-site health-services research. Yet methodological guidance for LLM integration into qualitative analysis and evidence of their impact on real-world research methods and outcomes remain limited. We developed a model- and task-agnostic framework for designing human-LLM qualitative analysis methods to support diverse analytic aims. Within a multi-site study of diabetes care at Federally Qualified Health Centers (FQHCs), we leveraged the framework to implement human-LLM methods for (1) qualitative synthesis of researcher-generated summaries to produce comparative feedback reports and (2) deductive coding of 167 interview transcripts to refine a practice-transformation intervention. LLM assistance enabled timely feedback to practitioners and the incorporation of large-scale qualitative data to inform theory and practice changes. This work demonstrates how LLMs can be integrated into applied health-services research to enhance efficiency while preserving rigor, offering guidance for continued innovation with LLMs in qualitative research.
Training-Free Adaptation of New-Generation LLMs using Legacy Clinical Models
Jan 06, 2026Abstract:Adapting language models to the clinical domain through continued pretraining and fine-tuning requires costly retraining for each new model generation. We propose Cross-Architecture Proxy Tuning (CAPT), a model-ensembling approach that enables training-free adaptation of state-of-the-art general-domain models using existing clinical models. CAPT supports models with disjoint vocabularies, leveraging contrastive decoding to selectively inject clinically relevant signals while preserving the general-domain model's reasoning and fluency. On six clinical classification and text-generation tasks, CAPT with a new-generation general-domain model and an older-generation clinical model consistently outperforms both models individually and state-of-the-art ensembling approaches (average +17.6% over UniTE, +41.4% over proxy tuning across tasks). Through token-level analysis and physician case studies, we demonstrate that CAPT amplifies clinically actionable language, reduces context errors, and increases clinical specificity.
Monitoring Deployed AI Systems in Health Care
Dec 09, 2025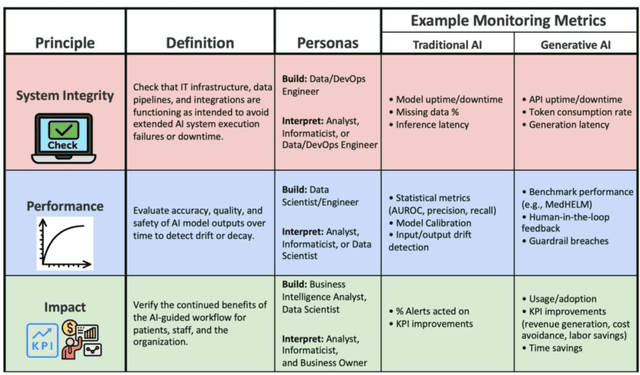
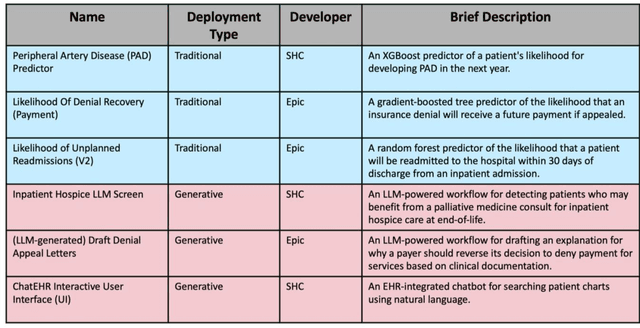
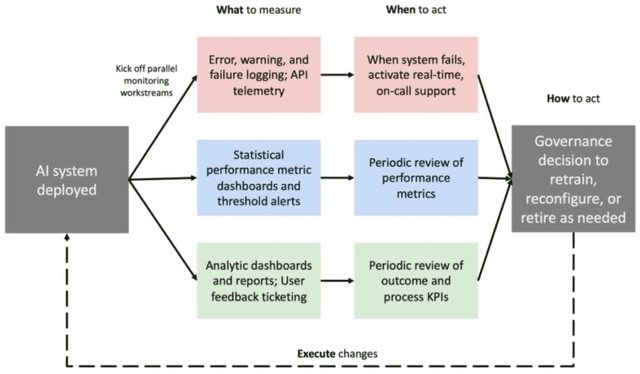
Abstract:Post-deployment monitoring of artificial intelligence (AI) systems in health care is essential to ensure their safety, quality, and sustained benefit-and to support governance decisions about which systems to update, modify, or decommission. Motivated by these needs, we developed a framework for monitoring deployed AI systems grounded in the mandate to take specific actions when they fail to behave as intended. This framework, which is now actively used at Stanford Health Care, is organized around three complementary principles: system integrity, performance, and impact. System integrity monitoring focuses on maximizing system uptime, detecting runtime errors, and identifying when changes to the surrounding IT ecosystem have unintended effects. Performance monitoring focuses on maintaining accurate system behavior in the face of changing health care practices (and thus input data) over time. Impact monitoring assesses whether a deployed system continues to have value in the form of benefit to clinicians and patients. Drawing on examples of deployed AI systems at our academic medical center, we provide practical guidance for creating monitoring plans based on these principles that specify which metrics to measure, when those metrics should be reviewed, who is responsible for acting when metrics change, and what concrete follow-up actions should be taken-for both traditional and generative AI. We also discuss challenges to implementing this framework, including the effort and cost of monitoring for health systems with limited resources and the difficulty of incorporating data-driven monitoring practices into complex organizations where conflicting priorities and definitions of success often coexist. This framework offers a practical template and starting point for health systems seeking to ensure that AI deployments remain safe and effective over time.
Retrieval-Augmented Guardrails for AI-Drafted Patient-Portal Messages: Error Taxonomy Construction and Large-Scale Evaluation
Sep 26, 2025Abstract:Asynchronous patient-clinician messaging via EHR portals is a growing source of clinician workload, prompting interest in large language models (LLMs) to assist with draft responses. However, LLM outputs may contain clinical inaccuracies, omissions, or tone mismatches, making robust evaluation essential. Our contributions are threefold: (1) we introduce a clinically grounded error ontology comprising 5 domains and 59 granular error codes, developed through inductive coding and expert adjudication; (2) we develop a retrieval-augmented evaluation pipeline (RAEC) that leverages semantically similar historical message-response pairs to improve judgment quality; and (3) we provide a two-stage prompting architecture using DSPy to enable scalable, interpretable, and hierarchical error detection. Our approach assesses the quality of drafts both in isolation and with reference to similar past message-response pairs retrieved from institutional archives. Using a two-stage DSPy pipeline, we compared baseline and reference-enhanced evaluations on over 1,500 patient messages. Retrieval context improved error identification in domains such as clinical completeness and workflow appropriateness. Human validation on 100 messages demonstrated superior agreement (concordance = 50% vs. 33%) and performance (F1 = 0.500 vs. 0.256) of context-enhanced labels vs. baseline, supporting the use of our RAEC pipeline as AI guardrails for patient messaging.
MedHELM: Holistic Evaluation of Large Language Models for Medical Tasks
May 26, 2025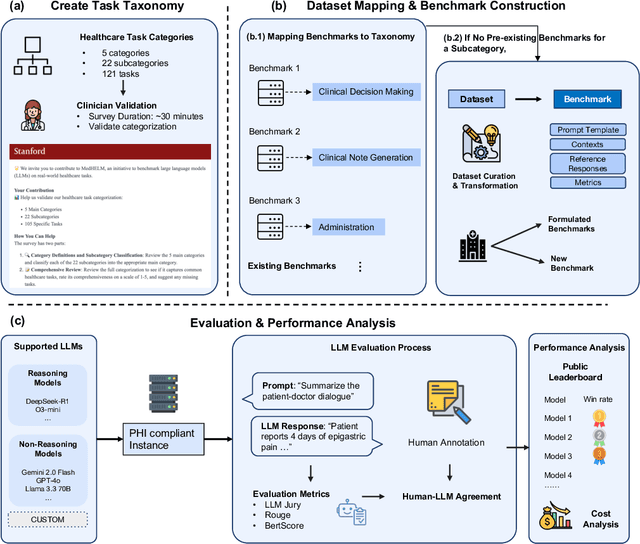
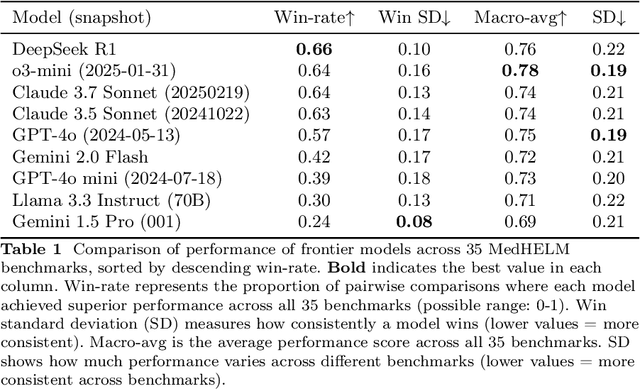
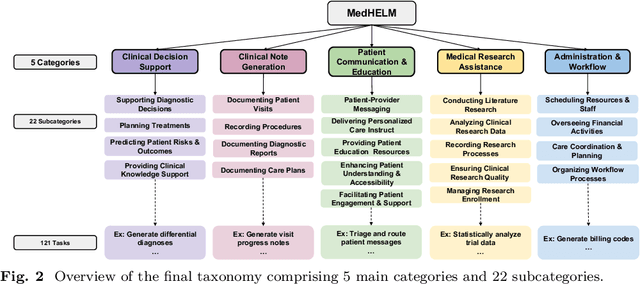
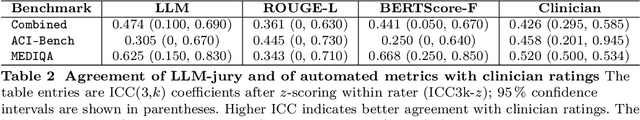
Abstract:While large language models (LLMs) achieve near-perfect scores on medical licensing exams, these evaluations inadequately reflect the complexity and diversity of real-world clinical practice. We introduce MedHELM, an extensible evaluation framework for assessing LLM performance for medical tasks with three key contributions. First, a clinician-validated taxonomy spanning 5 categories, 22 subcategories, and 121 tasks developed with 29 clinicians. Second, a comprehensive benchmark suite comprising 35 benchmarks (17 existing, 18 newly formulated) providing complete coverage of all categories and subcategories in the taxonomy. Third, a systematic comparison of LLMs with improved evaluation methods (using an LLM-jury) and a cost-performance analysis. Evaluation of 9 frontier LLMs, using the 35 benchmarks, revealed significant performance variation. Advanced reasoning models (DeepSeek R1: 66% win-rate; o3-mini: 64% win-rate) demonstrated superior performance, though Claude 3.5 Sonnet achieved comparable results at 40% lower estimated computational cost. On a normalized accuracy scale (0-1), most models performed strongly in Clinical Note Generation (0.73-0.85) and Patient Communication & Education (0.78-0.83), moderately in Medical Research Assistance (0.65-0.75), and generally lower in Clinical Decision Support (0.56-0.72) and Administration & Workflow (0.53-0.63). Our LLM-jury evaluation method achieved good agreement with clinician ratings (ICC = 0.47), surpassing both average clinician-clinician agreement (ICC = 0.43) and automated baselines including ROUGE-L (0.36) and BERTScore-F1 (0.44). Claude 3.5 Sonnet achieved comparable performance to top models at lower estimated cost. These findings highlight the importance of real-world, task-specific evaluation for medical use of LLMs and provides an open source framework to enable this.
BRIDGE: Benchmarking Large Language Models for Understanding Real-world Clinical Practice Text
May 01, 2025



Abstract:Large language models (LLMs) hold great promise for medical applications and are evolving rapidly, with new models being released at an accelerated pace. However, current evaluations of LLMs in clinical contexts remain limited. Most existing benchmarks rely on medical exam-style questions or PubMed-derived text, failing to capture the complexity of real-world electronic health record (EHR) data. Others focus narrowly on specific application scenarios, limiting their generalizability across broader clinical use. To address this gap, we present BRIDGE, a comprehensive multilingual benchmark comprising 87 tasks sourced from real-world clinical data sources across nine languages. We systematically evaluated 52 state-of-the-art LLMs (including DeepSeek-R1, GPT-4o, Gemini, and Llama 4) under various inference strategies. With a total of 13,572 experiments, our results reveal substantial performance variation across model sizes, languages, natural language processing tasks, and clinical specialties. Notably, we demonstrate that open-source LLMs can achieve performance comparable to proprietary models, while medically fine-tuned LLMs based on older architectures often underperform versus updated general-purpose models. The BRIDGE and its corresponding leaderboard serve as a foundational resource and a unique reference for the development and evaluation of new LLMs in real-world clinical text understanding. The BRIDGE leaderboard: https://huggingface.co/spaces/YLab-Open/BRIDGE-Medical-Leaderboard
TIMER: Temporal Instruction Modeling and Evaluation for Longitudinal Clinical Records
Mar 06, 2025Abstract:Large language models (LLMs) have emerged as promising tools for assisting in medical tasks, yet processing Electronic Health Records (EHRs) presents unique challenges due to their longitudinal nature. While LLMs' capabilities to perform medical tasks continue to improve, their ability to reason over temporal dependencies across multiple patient visits and time frames remains unexplored. We introduce TIMER (Temporal Instruction Modeling and Evaluation for Longitudinal Clinical Records), a framework that incorporate instruction-response pairs grounding to different parts of a patient's record as a critical dimension in both instruction evaluation and tuning for longitudinal clinical records. We develop TIMER-Bench, the first time-aware benchmark that evaluates temporal reasoning capabilities over longitudinal EHRs, as well as TIMER-Instruct, an instruction-tuning methodology for LLMs to learn reasoning over time. We demonstrate that models fine-tuned with TIMER-Instruct improve performance by 7.3% on human-generated benchmarks and 9.2% on TIMER-Bench, indicating that temporal instruction-tuning improves model performance for reasoning over EHR.
Identifying Reasons for Contraceptive Switching from Real-World Data Using Large Language Models
Feb 06, 2024Abstract:Prescription contraceptives play a critical role in supporting women's reproductive health. With nearly 50 million women in the United States using contraceptives, understanding the factors that drive contraceptives selection and switching is of significant interest. However, many factors related to medication switching are often only captured in unstructured clinical notes and can be difficult to extract. Here, we evaluate the zero-shot abilities of a recently developed large language model, GPT-4 (via HIPAA-compliant Microsoft Azure API), to identify reasons for switching between classes of contraceptives from the UCSF Information Commons clinical notes dataset. We demonstrate that GPT-4 can accurately extract reasons for contraceptive switching, outperforming baseline BERT-based models with microF1 scores of 0.849 and 0.881 for contraceptive start and stop extraction, respectively. Human evaluation of GPT-4-extracted reasons for switching showed 91.4% accuracy, with minimal hallucinations. Using extracted reasons, we identified patient preference, adverse events, and insurance as key reasons for switching using unsupervised topic modeling approaches. Notably, we also showed using our approach that "weight gain/mood change" and "insurance coverage" are disproportionately found as reasons for contraceptive switching in specific demographic populations. Our code and supplemental data are available at https://github.com/BMiao10/contraceptive-switching.
Do We Still Need Clinical Language Models?
Feb 16, 2023



Abstract:Although recent advances in scaling large language models (LLMs) have resulted in improvements on many NLP tasks, it remains unclear whether these models trained primarily with general web text are the right tool in highly specialized, safety critical domains such as clinical text. Recent results have suggested that LLMs encode a surprising amount of medical knowledge. This raises an important question regarding the utility of smaller domain-specific language models. With the success of general-domain LLMs, is there still a need for specialized clinical models? To investigate this question, we conduct an extensive empirical analysis of 12 language models, ranging from 220M to 175B parameters, measuring their performance on 3 different clinical tasks that test their ability to parse and reason over electronic health records. As part of our experiments, we train T5-Base and T5-Large models from scratch on clinical notes from MIMIC III and IV to directly investigate the efficiency of clinical tokens. We show that relatively small specialized clinical models substantially outperform all in-context learning approaches, even when finetuned on limited annotated data. Further, we find that pretraining on clinical tokens allows for smaller, more parameter-efficient models that either match or outperform much larger language models trained on general text. We release the code and the models used under the PhysioNet Credentialed Health Data license and data use agreement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge