Liyue Shen
Benchmarking Uncertainty Quantification of Plug-and-Play Diffusion Priors for Inverse Problems Solving
Feb 04, 2026Abstract:Plug-and-play diffusion priors (PnPDP) have become a powerful paradigm for solving inverse problems in scientific and engineering domains. Yet, current evaluations of reconstruction quality emphasize point-estimate accuracy metrics on a single sample, which do not reflect the stochastic nature of PnPDP solvers and the intrinsic uncertainty of inverse problems, critical for scientific tasks. This creates a fundamental mismatch: in inverse problems, the desired output is typically a posterior distribution and most PnPDP solvers induce a distribution over reconstructions, but existing benchmarks only evaluate a single reconstruction, ignoring distributional characterization such as uncertainty. To address this gap, we conduct a systematic study to benchmark the uncertainty quantification (UQ) of existing diffusion inverse solvers. Specifically, we design a rigorous toy model simulation to evaluate the uncertainty behavior of various PnPDP solvers, and propose a UQ-driven categorization. Through extensive experiments on toy simulations and diverse real-world scientific inverse problems, we observe uncertainty behaviors consistent with our taxonomy and theoretical justification, providing new insights for evaluating and understanding the uncertainty for PnPDPs.
Weak Diffusion Priors Can Still Achieve Strong Inverse-Problem Performance
Jan 30, 2026Abstract:Can a diffusion model trained on bedrooms recover human faces? Diffusion models are widely used as priors for inverse problems, but standard approaches usually assume a high-fidelity model trained on data that closely match the unknown signal. In practice, one often must use a mismatched or low-fidelity diffusion prior. Surprisingly, these weak priors often perform nearly as well as full-strength, in-domain baselines. We study when and why inverse solvers are robust to weak diffusion priors. Through extensive experiments, we find that weak priors succeed when measurements are highly informative (e.g., many observed pixels), and we identify regimes where they fail. Our theory, based on Bayesian consistency, gives conditions under which high-dimensional measurements make the posterior concentrate near the true signal. These results provide a principled justification on when weak diffusion priors can be used reliably.
Local Patches Meet Global Context: Scalable 3D Diffusion Priors for Computed Tomography Reconstruction
Dec 20, 2025



Abstract:Diffusion models learn strong image priors that can be leveraged to solve inverse problems like medical image reconstruction. However, for real-world applications such as 3D Computed Tomography (CT) imaging, directly training diffusion models on 3D data presents significant challenges due to the high computational demands of extensive GPU resources and large-scale datasets. Existing works mostly reuse 2D diffusion priors to address 3D inverse problems, but fail to fully realize and leverage the generative capacity of diffusion models for high-dimensional data. In this study, we propose a novel 3D patch-based diffusion model that can learn a fully 3D diffusion prior from limited data, enabling scalable generation of high-resolution 3D images. Our core idea is to learn the prior of 3D patches to achieve scalable efficiency, while coupling local and global information to guarantee high-quality 3D image generation, by modeling the joint distribution of position-aware 3D local patches and downsampled 3D volume as global context. Our approach not only enables high-quality 3D generation, but also offers an unprecedentedly efficient and accurate solution to high-resolution 3D inverse problems. Experiments on 3D CT reconstruction across multiple datasets show that our method outperforms state-of-the-art methods in both performance and efficiency, notably achieving high-resolution 3D reconstruction of $512 \times 512 \times 256$ ($\sim$20 mins).
Antithetic Noise in Diffusion Models
Jun 06, 2025Abstract:We initiate a systematic study of antithetic initial noise in diffusion models. Across unconditional models trained on diverse datasets, text-conditioned latent-diffusion models, and diffusion-posterior samplers, we find that pairing each initial noise with its negation consistently yields strongly negatively correlated samples. To explain this phenomenon, we combine experiments and theoretical analysis, leading to a symmetry conjecture that the learned score function is approximately affine antisymmetric (odd symmetry up to a constant shift), and provide evidence supporting it. Leveraging this negative correlation, we enable two applications: (1) enhancing image diversity in models like Stable Diffusion without quality loss, and (2) sharpening uncertainty quantification (e.g., up to 90% narrower confidence intervals) when estimating downstream statistics. Building on these gains, we extend the two-point pairing to a randomized quasi-Monte Carlo estimator, which further improves estimation accuracy. Our framework is training-free, model-agnostic, and adds no runtime overhead.
CCS: Controllable and Constrained Sampling with Diffusion Models via Initial Noise Perturbation
Feb 07, 2025



Abstract:Diffusion models have emerged as powerful tools for generative tasks, producing high-quality outputs across diverse domains. However, how the generated data responds to the initial noise perturbation in diffusion models remains under-explored, which hinders understanding the controllability of the sampling process. In this work, we first observe an interesting phenomenon: the relationship between the change of generation outputs and the scale of initial noise perturbation is highly linear through the diffusion ODE sampling. Then we provide both theoretical and empirical study to justify this linearity property of this input-output (noise-generation data) relationship. Inspired by these new insights, we propose a novel Controllable and Constrained Sampling method (CCS) together with a new controller algorithm for diffusion models to sample with desired statistical properties while preserving good sample quality. We perform extensive experiments to compare our proposed sampling approach with other methods on both sampling controllability and sampled data quality. Results show that our CCS method achieves more precisely controlled sampling while maintaining superior sample quality and diversity.
ST-NeRP: Spatial-Temporal Neural Representation Learning with Prior Embedding for Patient-specific Imaging Study
Oct 25, 2024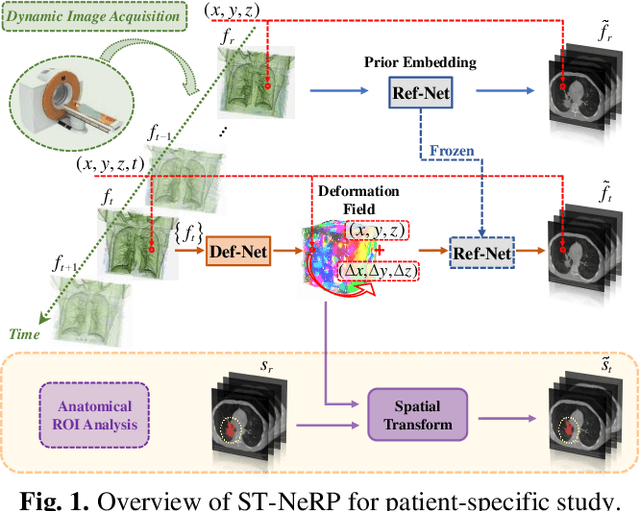
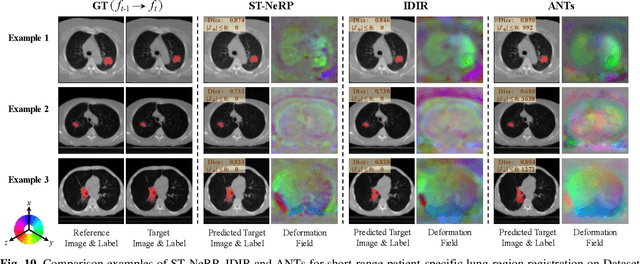
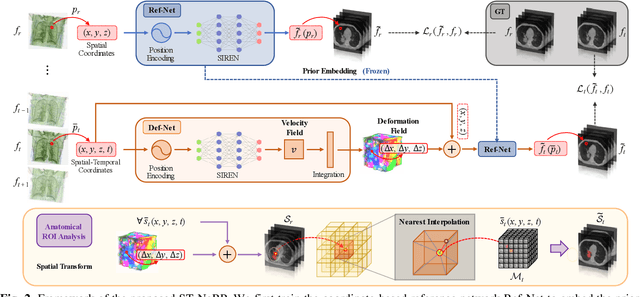
Abstract:During and after a course of therapy, imaging is routinely used to monitor the disease progression and assess the treatment responses. Despite of its significance, reliably capturing and predicting the spatial-temporal anatomic changes from a sequence of patient-specific image series presents a considerable challenge. Thus, the development of a computational framework becomes highly desirable for a multitude of practical applications. In this context, we propose a strategy of Spatial-Temporal Neural Representation learning with Prior embedding (ST-NeRP) for patient-specific imaging study. Our strategy involves leveraging an Implicit Neural Representation (INR) network to encode the image at the reference time point into a prior embedding. Subsequently, a spatial-temporally continuous deformation function is learned through another INR network. This network is trained using the whole patient-specific image sequence, enabling the prediction of deformation fields at various target time points. The efficacy of the ST-NeRP model is demonstrated through its application to diverse sequential image series, including 4D CT and longitudinal CT datasets within thoracic and abdominal imaging. The proposed ST-NeRP model exhibits substantial potential in enabling the monitoring of anatomical changes within a patient throughout the therapeutic journey.
Patch-Based Diffusion Models Beat Whole-Image Models for Mismatched Distribution Inverse Problems
Oct 15, 2024



Abstract:Diffusion models have achieved excellent success in solving inverse problems due to their ability to learn strong image priors, but existing approaches require a large training dataset of images that should come from the same distribution as the test dataset. When the training and test distributions are mismatched, artifacts and hallucinations can occur in reconstructed images due to the incorrect priors. In this work, we systematically study out of distribution (OOD) problems where a known training distribution is first provided. We first study the setting where only a single measurement obtained from the unknown test distribution is available. Next we study the setting where a very small sample of data belonging to the test distribution is available, and our goal is still to reconstruct an image from a measurement that came from the test distribution. In both settings, we use a patch-based diffusion prior that learns the image distribution solely from patches. Furthermore, in the first setting, we include a self-supervised loss that helps the network output maintain consistency with the measurement. Extensive experiments show that in both settings, the patch-based method can obtain high quality image reconstructions that can outperform whole-image models and can compete with methods that have access to large in-distribution training datasets. Furthermore, we show how whole-image models are prone to memorization and overfitting, leading to artifacts in the reconstructions, while a patch-based model can resolve these issues.
Latent Space Disentanglement in Diffusion Transformers Enables Zero-shot Fine-grained Semantic Editing
Aug 23, 2024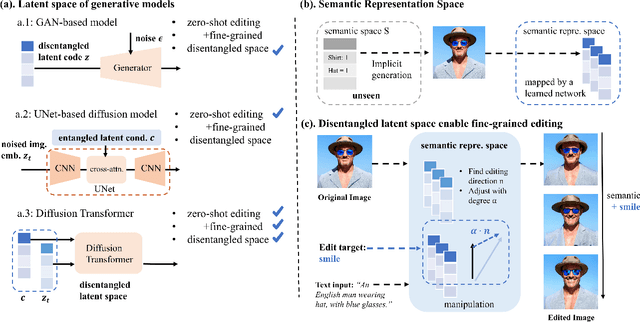

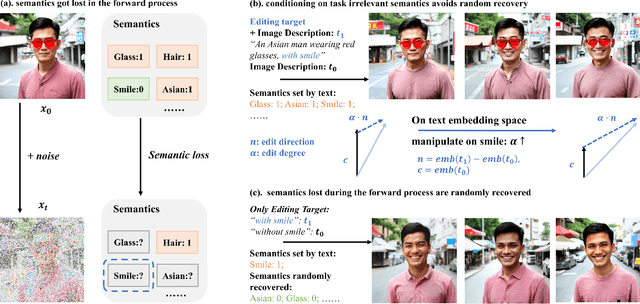

Abstract:Diffusion Transformers (DiTs) have achieved remarkable success in diverse and high-quality text-to-image(T2I) generation. However, how text and image latents individually and jointly contribute to the semantics of generated images, remain largely unexplored. Through our investigation of DiT's latent space, we have uncovered key findings that unlock the potential for zero-shot fine-grained semantic editing: (1) Both the text and image spaces in DiTs are inherently decomposable. (2) These spaces collectively form a disentangled semantic representation space, enabling precise and fine-grained semantic control. (3) Effective image editing requires the combined use of both text and image latent spaces. Leveraging these insights, we propose a simple and effective Extract-Manipulate-Sample (EMS) framework for zero-shot fine-grained image editing. Our approach first utilizes a multi-modal Large Language Model to convert input images and editing targets into text descriptions. We then linearly manipulate text embeddings based on the desired editing degree and employ constrained score distillation sampling to manipulate image embeddings. We quantify the disentanglement degree of the latent space of diffusion models by proposing a new metric. To evaluate fine-grained editing performance, we introduce a comprehensive benchmark incorporating both human annotations, manual evaluation, and automatic metrics. We have conducted extensive experimental results and in-depth analysis to thoroughly uncover the semantic disentanglement properties of the diffusion transformer, as well as the effectiveness of our proposed method. Our annotated benchmark dataset is publicly available at https://anonymous.com/anonymous/EMS-Benchmark, facilitating reproducible research in this domain.
CoSIGN: Few-Step Guidance of ConSIstency Model to Solve General INverse Problems
Jul 17, 2024Abstract:Diffusion models have been demonstrated as strong priors for solving general inverse problems. Most existing Diffusion model-based Inverse Problem Solvers (DIS) employ a plug-and-play approach to guide the sampling trajectory with either projections or gradients. Though effective, these methods generally necessitate hundreds of sampling steps, posing a dilemma between inference time and reconstruction quality. In this work, we try to push the boundary of inference steps to 1-2 NFEs while still maintaining high reconstruction quality. To achieve this, we propose to leverage a pretrained distillation of diffusion model, namely consistency model, as the data prior. The key to achieving few-step guidance is to enforce two types of constraints during the sampling process of the consistency model: soft measurement constraint with ControlNet and hard measurement constraint via optimization. Supporting both single-step reconstruction and multistep refinement, the proposed framework further provides a way to trade image quality with additional computational cost. Within comparable NFEs, our method achieves new state-of-the-art in diffusion-based inverse problem solving, showcasing the significant potential of employing prior-based inverse problem solvers for real-world applications. Code is available at: https://github.com/BioMed-AI-Lab-U-Michgan/cosign.
Efficient In-Context Medical Segmentation with Meta-driven Visual Prompt Selection
Jul 15, 2024



Abstract:In-context learning (ICL) with Large Vision Models (LVMs) presents a promising avenue in medical image segmentation by reducing the reliance on extensive labeling. However, the ICL performance of LVMs highly depends on the choices of visual prompts and suffers from domain shifts. While existing works leveraging LVMs for medical tasks have focused mainly on model-centric approaches like fine-tuning, we study an orthogonal data-centric perspective on how to select good visual prompts to facilitate generalization to medical domain. In this work, we propose a label-efficient in-context medical segmentation method by introducing a novel Meta-driven Visual Prompt Selection mechanism (MVPS), where a prompt retriever obtained from a meta-learning framework actively selects the optimal images as prompts to promote model performance and generalizability. Evaluated on 8 datasets and 4 tasks across 3 medical imaging modalities, our proposed approach demonstrates consistent gains over existing methods under different scenarios, improving both computational and label efficiency. Finally, we show that MVPS is a flexible, finetuning-free module that could be easily plugged into different backbones and combined with other model-centric approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge