Jason Hu
Local Patches Meet Global Context: Scalable 3D Diffusion Priors for Computed Tomography Reconstruction
Dec 20, 2025



Abstract:Diffusion models learn strong image priors that can be leveraged to solve inverse problems like medical image reconstruction. However, for real-world applications such as 3D Computed Tomography (CT) imaging, directly training diffusion models on 3D data presents significant challenges due to the high computational demands of extensive GPU resources and large-scale datasets. Existing works mostly reuse 2D diffusion priors to address 3D inverse problems, but fail to fully realize and leverage the generative capacity of diffusion models for high-dimensional data. In this study, we propose a novel 3D patch-based diffusion model that can learn a fully 3D diffusion prior from limited data, enabling scalable generation of high-resolution 3D images. Our core idea is to learn the prior of 3D patches to achieve scalable efficiency, while coupling local and global information to guarantee high-quality 3D image generation, by modeling the joint distribution of position-aware 3D local patches and downsampled 3D volume as global context. Our approach not only enables high-quality 3D generation, but also offers an unprecedentedly efficient and accurate solution to high-resolution 3D inverse problems. Experiments on 3D CT reconstruction across multiple datasets show that our method outperforms state-of-the-art methods in both performance and efficiency, notably achieving high-resolution 3D reconstruction of $512 \times 512 \times 256$ ($\sim$20 mins).
CCS: Controllable and Constrained Sampling with Diffusion Models via Initial Noise Perturbation
Feb 07, 2025



Abstract:Diffusion models have emerged as powerful tools for generative tasks, producing high-quality outputs across diverse domains. However, how the generated data responds to the initial noise perturbation in diffusion models remains under-explored, which hinders understanding the controllability of the sampling process. In this work, we first observe an interesting phenomenon: the relationship between the change of generation outputs and the scale of initial noise perturbation is highly linear through the diffusion ODE sampling. Then we provide both theoretical and empirical study to justify this linearity property of this input-output (noise-generation data) relationship. Inspired by these new insights, we propose a novel Controllable and Constrained Sampling method (CCS) together with a new controller algorithm for diffusion models to sample with desired statistical properties while preserving good sample quality. We perform extensive experiments to compare our proposed sampling approach with other methods on both sampling controllability and sampled data quality. Results show that our CCS method achieves more precisely controlled sampling while maintaining superior sample quality and diversity.
On Adapting Randomized Nyström Preconditioners to Accelerate Variational Image Reconstruction
Nov 12, 2024
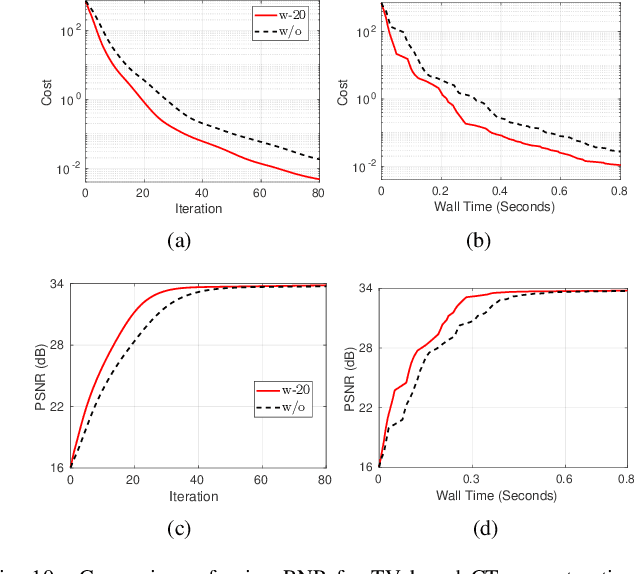
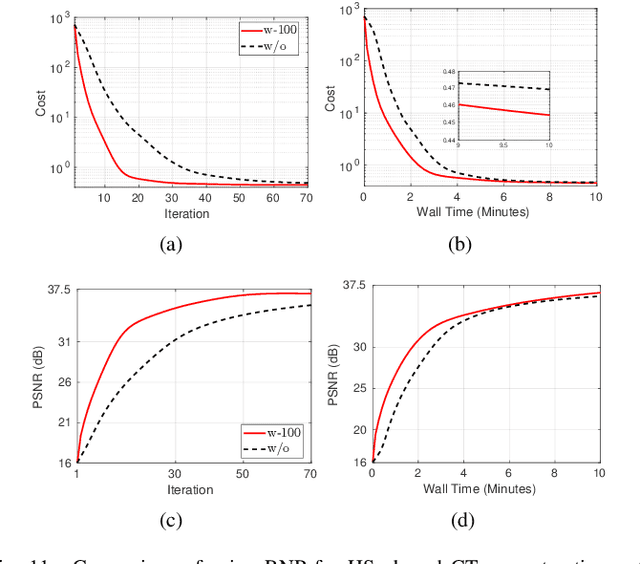
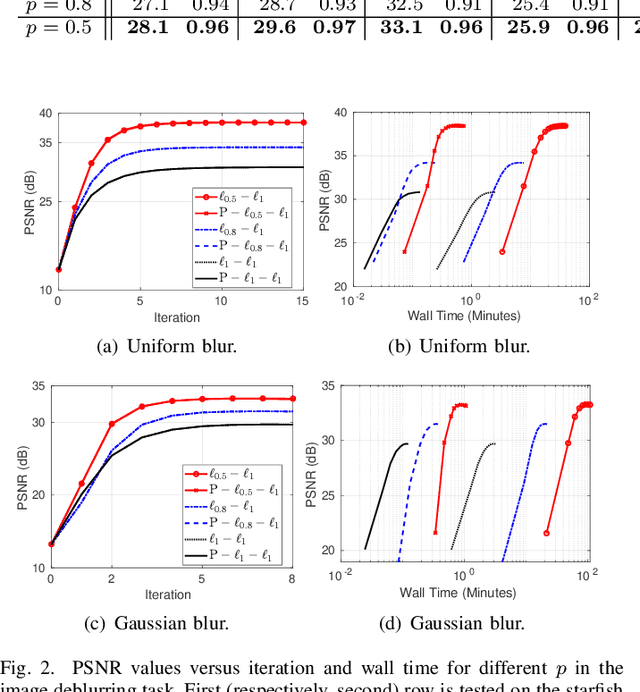
Abstract:Model-based iterative reconstruction plays a key role in solving inverse problems. However, the associated minimization problems are generally large-scale, ill-posed, nonsmooth, and sometimes even nonconvex, which present challenges in designing efficient iterative solvers and often prevent their practical use. Preconditioning methods can significantly accelerate the convergence of iterative methods. In some applications, computing preconditioners on-the-fly is beneficial. Moreover, forward models in image reconstruction are typically represented as operators, and the corresponding explicit matrices are often unavailable, which brings additional challenges in designing preconditioners. Therefore, for practical use, computing and applying preconditioners should be computationally inexpensive. This paper adapts the randomized Nystr\"{o}m approximation to compute effective preconditioners that accelerate image reconstruction without requiring an explicit matrix for the forward model. We leverage modern GPU computational platforms to compute the preconditioner on-the-fly. Moreover, we propose efficient approaches for applying the preconditioner to problems with nonsmooth regularizers. Our numerical results on image deblurring, super-resolution with impulsive noise, and computed tomography reconstruction demonstrate the efficiency and effectiveness of the proposed preconditioner.
Patch-Based Diffusion Models Beat Whole-Image Models for Mismatched Distribution Inverse Problems
Oct 15, 2024



Abstract:Diffusion models have achieved excellent success in solving inverse problems due to their ability to learn strong image priors, but existing approaches require a large training dataset of images that should come from the same distribution as the test dataset. When the training and test distributions are mismatched, artifacts and hallucinations can occur in reconstructed images due to the incorrect priors. In this work, we systematically study out of distribution (OOD) problems where a known training distribution is first provided. We first study the setting where only a single measurement obtained from the unknown test distribution is available. Next we study the setting where a very small sample of data belonging to the test distribution is available, and our goal is still to reconstruct an image from a measurement that came from the test distribution. In both settings, we use a patch-based diffusion prior that learns the image distribution solely from patches. Furthermore, in the first setting, we include a self-supervised loss that helps the network output maintain consistency with the measurement. Extensive experiments show that in both settings, the patch-based method can obtain high quality image reconstructions that can outperform whole-image models and can compete with methods that have access to large in-distribution training datasets. Furthermore, we show how whole-image models are prone to memorization and overfitting, leading to artifacts in the reconstructions, while a patch-based model can resolve these issues.
Shorter SPECT Scans Using Self-supervised Coordinate Learning to Synthesize Skipped Projection Views
Jun 27, 2024



Abstract:Purpose: This study addresses the challenge of extended SPECT imaging duration under low-count conditions, as encountered in Lu-177 SPECT imaging, by developing a self-supervised learning approach to synthesize skipped SPECT projection views, thus shortening scan times in clinical settings. Methods: We employed a self-supervised coordinate-based learning technique, adapting the neural radiance field (NeRF) concept in computer vision to synthesize under-sampled SPECT projection views. For each single scan, we used self-supervised coordinate learning to estimate skipped SPECT projection views. The method was tested with various down-sampling factors (DFs=2, 4, 8) on both Lu-177 phantom SPECT/CT measurements and clinical SPECT/CT datasets, from 11 patients undergoing Lu-177 DOTATATE and 6 patients undergoing Lu-177 PSMA-617 radiopharmaceutical therapy. Results: For SPECT reconstructions, our method outperformed the use of linearly interpolated projections and partial projection views in relative contrast-to-noise-ratios (RCNR) averaged across different downsampling factors: 1) DOTATATE: 83% vs. 65% vs. 67% for lesions and 86% vs. 70% vs. 67% for kidney, 2) PSMA: 76% vs. 69% vs. 68% for lesions and 75% vs. 55% vs. 66% for organs, including kidneys, lacrimal glands, parotid glands, and submandibular glands. Conclusion: The proposed method enables reduction in acquisition time (by factors of 2, 4, or 8) while maintaining quantitative accuracy in clinical SPECT protocols by allowing for the collection of fewer projections. Importantly, the self-supervised nature of this NeRF-based approach eliminates the need for extensive training data, instead learning from each patient's projection data alone. The reduction in acquisition time is particularly relevant for imaging under low-count conditions and for protocols that require multiple-bed positions such as whole-body imaging.
DiffusionBlend: Learning 3D Image Prior through Position-aware Diffusion Score Blending for 3D Computed Tomography Reconstruction
Jun 14, 2024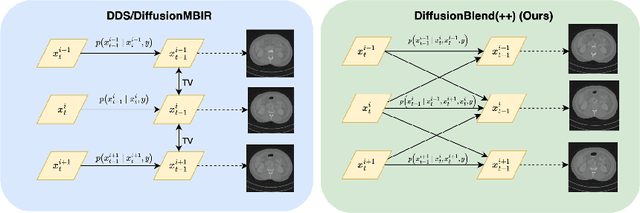
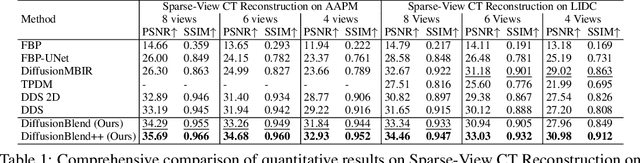
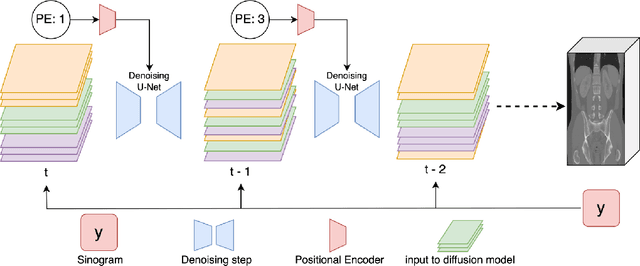
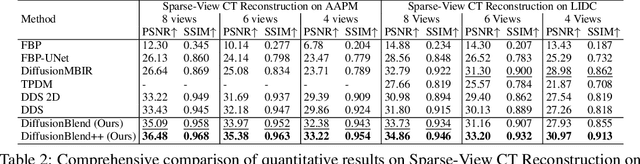
Abstract:Diffusion models face significant challenges when employed for large-scale medical image reconstruction in real practice such as 3D Computed Tomography (CT). Due to the demanding memory, time, and data requirements, it is difficult to train a diffusion model directly on the entire volume of high-dimensional data to obtain an efficient 3D diffusion prior. Existing works utilizing diffusion priors on single 2D image slice with hand-crafted cross-slice regularization would sacrifice the z-axis consistency, which results in severe artifacts along the z-axis. In this work, we propose a novel framework that enables learning the 3D image prior through position-aware 3D-patch diffusion score blending for reconstructing large-scale 3D medical images. To the best of our knowledge, we are the first to utilize a 3D-patch diffusion prior for 3D medical image reconstruction. Extensive experiments on sparse view and limited angle CT reconstruction show that our DiffusionBlend method significantly outperforms previous methods and achieves state-of-the-art performance on real-world CT reconstruction problems with high-dimensional 3D image (i.e., $256 \times 256 \times 500$). Our algorithm also comes with better or comparable computational efficiency than previous state-of-the-art methods.
Learning Image Priors through Patch-based Diffusion Models for Solving Inverse Problems
Jun 04, 2024



Abstract:Diffusion models can learn strong image priors from underlying data distribution and use them to solve inverse problems, but the training process is computationally expensive and requires lots of data. Such bottlenecks prevent most existing works from being feasible for high-dimensional and high-resolution data such as 3D images. This paper proposes a method to learn an efficient data prior for the entire image by training diffusion models only on patches of images. Specifically, we propose a patch-based position-aware diffusion inverse solver, called PaDIS, where we obtain the score function of the whole image through scores of patches and their positional encoding and utilize this as the prior for solving inverse problems. First of all, we show that this diffusion model achieves an improved memory efficiency and data efficiency while still maintaining the capability to generate entire images via positional encoding. Additionally, the proposed PaDIS model is highly flexible and can be plugged in with different diffusion inverse solvers (DIS). We demonstrate that the proposed PaDIS approach enables solving various inverse problems in both natural and medical image domains, including CT reconstruction, deblurring, and superresolution, given only patch-based priors. Notably, PaDIS outperforms previous DIS methods trained on entire image priors in the case of limited training data, demonstrating the data efficiency of our proposed approach by learning patch-based prior.
Provable Preconditioned Plug-and-Play Approach for Compressed Sensing MRI Reconstruction
May 06, 2024



Abstract:Model-based methods play a key role in the reconstruction of compressed sensing (CS) MRI. Finding an effective prior to describe the statistical distribution of the image family of interest is crucial for model-based methods. Plug-and-play (PnP) is a general framework that uses denoising algorithms as the prior or regularizer. Recent work showed that PnP methods with denoisers based on pretrained convolutional neural networks outperform other classical regularizers in CS MRI reconstruction. However, the numerical solvers for PnP can be slow for CS MRI reconstruction. This paper proposes a preconditioned PnP (P^2nP) method to accelerate the convergence speed. Moreover, we provide proofs of the fixed-point convergence of the P^2nP iterates. Numerical experiments on CS MRI reconstruction with non-Cartesian sampling trajectories illustrate the effectiveness and efficiency of the P^2nP approach.
AWFSD: Accelerated Wirtinger Flow with Score-based Diffusion Image Prior for Poisson-Gaussian Holographic Phase Retrieval
May 12, 2023



Abstract:Phase retrieval (PR) is an essential problem in a number of coherent imaging systems. This work aims at resolving the holographic phase retrieval problem in real world scenarios where the measurements are corrupted by a mixture of Poisson and Gaussian (PG) noise that stems from optical imaging systems. To solve this problem, we develop a novel algorithm based on Accelerated Wirtinger Flow that uses Score-based Diffusion models as the generative prior (AWFSD). In particular, we frame the PR problem as an optimization task that involves both a data fidelity term and a regularization term. We derive the gradient of the PG log-likelihood function along with its corresponding Lipschitz constant, ensuring a more accurate data consistency term for practical measurements. We introduce a generative prior as part of our regularization approach by using a score-based diffusion model to capture (the gradient of) the image prior distribution. We provide theoretical analysis that establishes a critical-point convergence guarantee for the proposed AWFSD algorithm. Our simulation experiments demonstrate that: 1) The proposed algorithm based on the PG likelihood model enhances reconstruction compared to that solely based on either Gaussian or Poisson likelihood. 2) The proposed AWFSD algorithm produces reconstructions with higher image quality both qualitatively and quantitatively, and is more robust to variations in noise levels when compared with state-of-the-art methods for phase retrieval.
Integrating cross-modality hallucinated MRI with CT to aid mediastinal lung tumor segmentation
Sep 10, 2019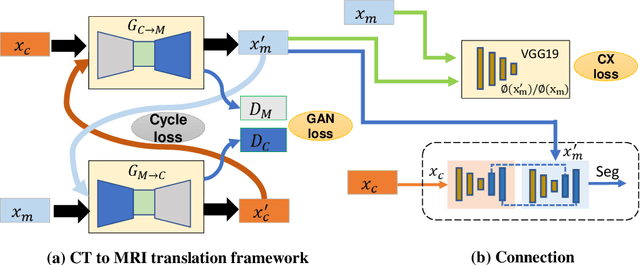
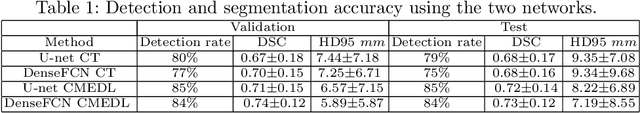
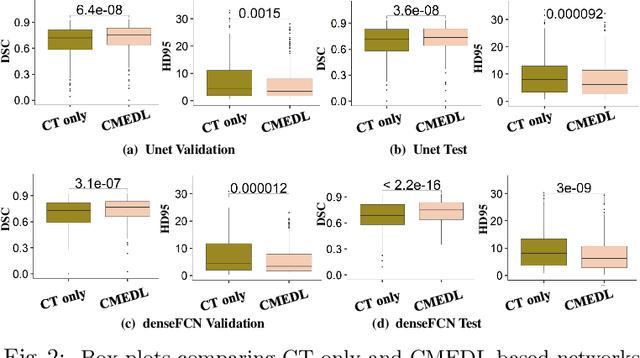
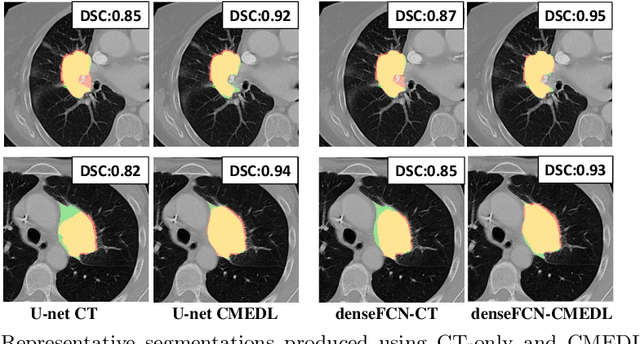
Abstract:Lung tumors, especially those located close to or surrounded by soft tissues like the mediastinum, are difficult to segment due to the low soft tissue contrast on computed tomography images. Magnetic resonance images contain superior soft-tissue contrast information that can be leveraged if both modalities were available for training. Therefore, we developed a cross-modality educed learning approach where MR information that is educed from CT is used to hallucinate MRI and improve CT segmentation. Our approach, called cross-modality educed deep learning segmentation (CMEDL) combines CT and pseudo MR produced from CT by aligning their features to obtain segmentation on CT. Features computed in the last two layers of parallelly trained CT and MR segmentation networks are aligned. We implemented this approach on U-net and dense fully convolutional networks (dense-FCN). Our networks were trained on unrelated cohorts from open-source the Cancer Imaging Archive CT images (N=377), an internal archive T2-weighted MR (N=81), and evaluated using separate validation (N=304) and testing (N=333) CT-delineated tumors. Our approach using both networks were significantly more accurate (U-net $P <0.001$; denseFCN $P <0.001$) than CT-only networks and achieved an accuracy (Dice similarity coefficient) of 0.71$\pm$0.15 (U-net), 0.74$\pm$0.12 (denseFCN) on validation and 0.72$\pm$0.14 (U-net), 0.73$\pm$0.12 (denseFCN) on the testing sets. Our novel approach demonstrated that educing cross-modality information through learned priors enhances CT segmentation performance
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge