Sean L. Berry
Nested-block self-attention for robust radiotherapy planning segmentation
Feb 26, 2021



Abstract:Although deep convolutional networks have been widely studied for head and neck (HN) organs at risk (OAR) segmentation, their use for routine clinical treatment planning is limited by a lack of robustness to imaging artifacts, low soft tissue contrast on CT, and the presence of abnormal anatomy. In order to address these challenges, we developed a computationally efficient nested block self-attention (NBSA) method that can be combined with any convolutional network. Our method achieves computational efficiency by performing non-local calculations within memory blocks of fixed spatial extent. Contextual dependencies are captured by passing information in a raster scan order between blocks, as well as through a second attention layer that causes bi-directional attention flow. We implemented our approach on three different networks to demonstrate feasibility. Following training using 200 cases, we performed comprehensive evaluations using conventional and clinical metrics on a separate set of 172 test scans sourced from external and internal institution datasets without any exclusion criteria. NBSA required a similar number of computations (15.7 gflops) as the most efficient criss-cross attention (CCA) method and generated significantly more accurate segmentations for brain stem (Dice of 0.89 vs. 0.86) and parotid glands (0.86 vs. 0.84) than CCA. NBSA's segmentations were less variable than multiple 3D methods, including for small organs with low soft-tissue contrast such as the submandibular glands (surface Dice of 0.90).
Integrating cross-modality hallucinated MRI with CT to aid mediastinal lung tumor segmentation
Sep 10, 2019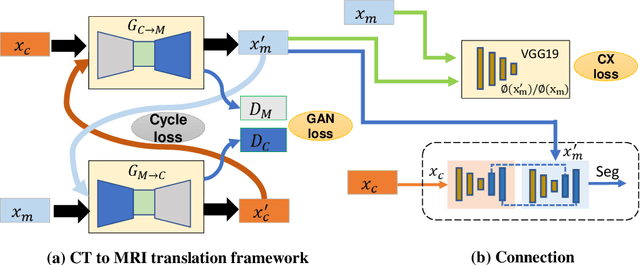
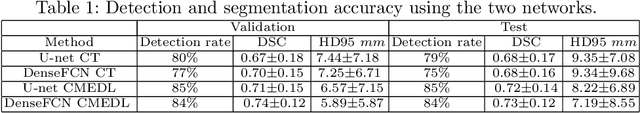
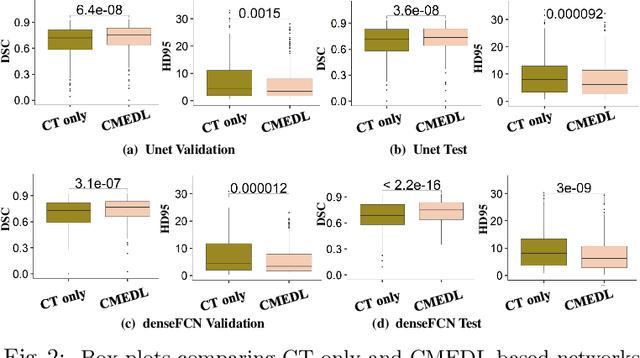
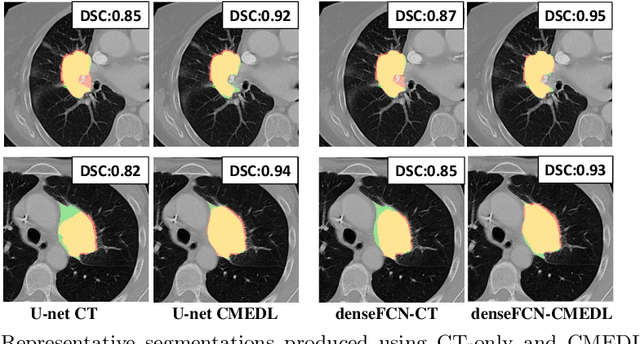
Abstract:Lung tumors, especially those located close to or surrounded by soft tissues like the mediastinum, are difficult to segment due to the low soft tissue contrast on computed tomography images. Magnetic resonance images contain superior soft-tissue contrast information that can be leveraged if both modalities were available for training. Therefore, we developed a cross-modality educed learning approach where MR information that is educed from CT is used to hallucinate MRI and improve CT segmentation. Our approach, called cross-modality educed deep learning segmentation (CMEDL) combines CT and pseudo MR produced from CT by aligning their features to obtain segmentation on CT. Features computed in the last two layers of parallelly trained CT and MR segmentation networks are aligned. We implemented this approach on U-net and dense fully convolutional networks (dense-FCN). Our networks were trained on unrelated cohorts from open-source the Cancer Imaging Archive CT images (N=377), an internal archive T2-weighted MR (N=81), and evaluated using separate validation (N=304) and testing (N=333) CT-delineated tumors. Our approach using both networks were significantly more accurate (U-net $P <0.001$; denseFCN $P <0.001$) than CT-only networks and achieved an accuracy (Dice similarity coefficient) of 0.71$\pm$0.15 (U-net), 0.74$\pm$0.12 (denseFCN) on validation and 0.72$\pm$0.14 (U-net), 0.73$\pm$0.12 (denseFCN) on the testing sets. Our novel approach demonstrated that educing cross-modality information through learned priors enhances CT segmentation performance
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge