Neelam Tyagi
Segmentation Regularized Training for Multi-Domain Deep Learning Registration applied to MR-Guided Prostate Cancer Radiotherapy
Jul 09, 2025Abstract:Background: Accurate deformable image registration (DIR) is required for contour propagation and dose accumulation in MR-guided adaptive radiotherapy (MRgART). This study trained and evaluated a deep learning DIR method for domain invariant MR-MR registration. Methods: A progressively refined registration and segmentation (ProRSeg) method was trained with 262 pairs of 3T MR simulation scans from prostate cancer patients using weighted segmentation consistency loss. ProRSeg was tested on same- (58 pairs), cross- (72 1.5T MR Linac pairs), and mixed-domain (42 MRSim-MRL pairs) datasets for contour propagation accuracy of clinical target volume (CTV), bladder, and rectum. Dose accumulation was performed for 42 patients undergoing 5-fraction MRgART. Results: ProRSeg demonstrated generalization for bladder with similar Dice Similarity Coefficients across domains (0.88, 0.87, 0.86). For rectum and CTV, performance was domain-dependent with higher accuracy on cross-domain MRL dataset (DSCs 0.89) versus same-domain data. The model's strong cross-domain performance prompted us to study the feasibility of using it for dose accumulation. Dose accumulation showed 83.3% of patients met CTV coverage (D95 >= 40.0 Gy) and bladder sparing (D50 <= 20.0 Gy) constraints. All patients achieved minimum mean target dose (>40.4 Gy), but only 9.5% remained under upper limit (<42.0 Gy). Conclusions: ProRSeg showed reasonable multi-domain MR-MR registration performance for prostate cancer patients with preliminary feasibility for evaluating treatment compliance to clinical constraints.
Modality-agnostic, patient-specific digital twins modeling temporally varying digestive motion
Jul 03, 2025Abstract:Objective: Clinical implementation of deformable image registration (DIR) requires voxel-based spatial accuracy metrics such as manually identified landmarks, which are challenging to implement for highly mobile gastrointestinal (GI) organs. To address this, patient-specific digital twins (DT) modeling temporally varying motion were created to assess the accuracy of DIR methods. Approach: 21 motion phases simulating digestive GI motion as 4D sequences were generated from static 3D patient scans using published analytical GI motion models through a semi-automated pipeline. Eleven datasets, including six T2w FSE MRI (T2w MRI), two T1w 4D golden-angle stack-of-stars, and three contrast-enhanced CT scans. The motion amplitudes of the DTs were assessed against real patient stomach motion amplitudes extracted from independent 4D MRI datasets. The generated DTs were then used to assess six different DIR methods using target registration error, Dice similarity coefficient, and the 95th percentile Hausdorff distance using summary metrics and voxel-level granular visualizations. Finally, for a subset of T2w MRI scans from patients treated with MR-guided radiation therapy, dose distributions were warped and accumulated to assess dose warping errors, including evaluations of DIR performance in both low- and high-dose regions for patient-specific error estimation. Main results: Our proposed pipeline synthesized DTs modeling realistic GI motion, achieving mean and maximum motion amplitudes and a mean log Jacobian determinant within 0.8 mm and 0.01, respectively, similar to published real-patient gastric motion data. It also enables the extraction of detailed quantitative DIR performance metrics and rigorous validation of dose mapping accuracy. Significance: The pipeline enables rigorously testing DIR tools for dynamic, anatomically complex regions enabling granular spatial and dosimetric accuracies.
Deep Learning Based Dominant Index Lesion Segmentation for MR-guided Radiation Therapy of Prostate Cancer
Mar 06, 2023Abstract:Dose escalation radiotherapy allows increased control of prostate cancer (PCa) but requires segmentation of dominant index lesions (DIL), motivating the development of automated methods for fast, accurate, and consistent segmentation of PCa DIL. We evaluated five deep-learning networks on apparent diffusion coefficient (ADC) MRI from 500 lesions in 365 patients arising from internal training Dataset 1 (1.5Tesla GE MR with endorectal coil), external ProstateX Dataset 2 (3Tesla Siemens MR), and internal inter-rater Dataset 3 (3Tesla Philips MR). The networks include: multiple resolution residually connected network (MRRN) and MRRN regularized in training with deep supervision (MRRN-DS), Unet, Unet++, ResUnet, and fast panoptic segmentation (FPSnet) as well as fast panoptic segmentation with smoothed labels (FPSnet-SL). Models were evaluated by volumetric DIL segmentation accuracy using Dice similarity coefficient (DSC) and detection accuracy, as a function of lesion aggressiveness, size, and location (Dataset 1 and 2), and accuracy with respect to two-raters (on Dataset 3). In general MRRN-DS more accurately segmented tumors than other methods on the testing datasets. MRRN-DS significantly outperformed ResUnet in Dataset2 (DSC of 0.54 vs. 0.44, p<0.001) and the Unet++ in Dataset3 (DSC of 0.45 vs. p=0.04). FPSnet-SL was similarly accurate as MRRN-DS in Dataset2 (p = 0.30), but MRRN-DS significantly outperformed FPSnet and FPSnet-SL in both Dataset1 (0.60 vs 0.51 [p=0.01] and 0.54 [p=0.049] respectively) and Dataset3 (0.45 vs 0.06 [p=0.002] and 0.24 [p=0.004] respectively). Finally, MRRN-DS produced slightly higher agreement with experienced radiologist than two radiologists in Dataset 3 (DSC of 0.45 vs. 0.41).
* https://pubmed.ncbi.nlm.nih.gov/36856092/
Progressively refined deep joint registration segmentation (ProRSeg) of gastrointestinal organs at risk: Application to MRI and cone-beam CT
Oct 25, 2022



Abstract:Method: ProRSeg was trained using 5-fold cross-validation with 110 T2-weighted MRI acquired at 5 treatment fractions from 10 different patients, taking care that same patient scans were not placed in training and testing folds. Segmentation accuracy was measured using Dice similarity coefficient (DSC) and Hausdorff distance at 95th percentile (HD95). Registration consistency was measured using coefficient of variation (CV) in displacement of OARs. Ablation tests and accuracy comparisons against multiple methods were done. Finally, applicability of ProRSeg to segment cone-beam CT (CBCT) scans was evaluated on 80 scans using 5-fold cross-validation. Results: ProRSeg processed 3D volumes (128 $\times$ 192 $\times$ 128) in 3 secs on a NVIDIA Tesla V100 GPU. It's segmentations were significantly more accurate ($p<0.001$) than compared methods, achieving a DSC of 0.94 $\pm$0.02 for liver, 0.88$\pm$0.04 for large bowel, 0.78$\pm$0.03 for small bowel and 0.82$\pm$0.04 for stomach-duodenum from MRI. ProRSeg achieved a DSC of 0.72$\pm$0.01 for small bowel and 0.76$\pm$0.03 for stomach-duodenum from CBCT. ProRSeg registrations resulted in the lowest CV in displacement (stomach-duodenum $CV_{x}$: 0.75\%, $CV_{y}$: 0.73\%, and $CV_{z}$: 0.81\%; small bowel $CV_{x}$: 0.80\%, $CV_{y}$: 0.80\%, and $CV_{z}$: 0.68\%; large bowel $CV_{x}$: 0.71\%, $CV_{y}$ : 0.81\%, and $CV_{z}$: 0.75\%). ProRSeg based dose accumulation accounting for intra-fraction (pre-treatment to post-treatment MRI scan) and inter-fraction motion showed that the organ dose constraints were violated in 4 patients for stomach-duodenum and for 3 patients for small bowel. Study limitations include lack of independent testing and ground truth phantom datasets to measure dose accumulation accuracy.
Self-supervised 3D anatomy segmentation using self-distilled masked image transformer (SMIT)
May 20, 2022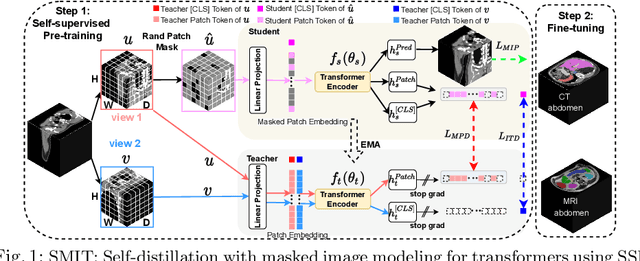
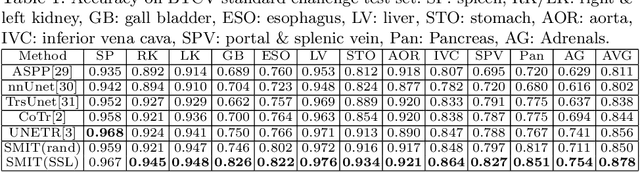
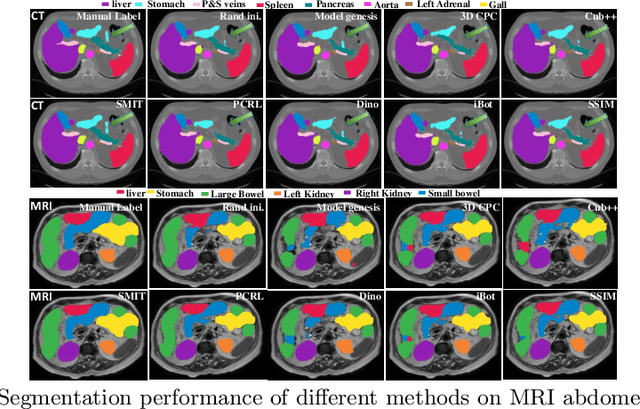
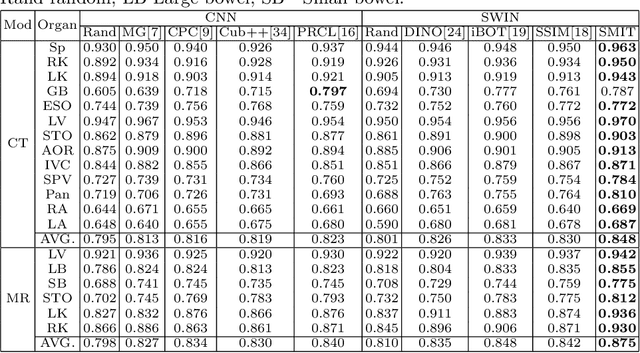
Abstract:Vision transformers, with their ability to more efficiently model long-range context, have demonstrated impressive accuracy gains in several computer vision and medical image analysis tasks including segmentation. However, such methods need large labeled datasets for training, which is hard to obtain for medical image analysis. Self-supervised learning (SSL) has demonstrated success in medical image segmentation using convolutional networks. In this work, we developed a \underline{s}elf-distillation learning with \underline{m}asked \underline{i}mage modeling method to perform SSL for vision \underline{t}ransformers (SMIT) applied to 3D multi-organ segmentation from CT and MRI. Our contribution is a dense pixel-wise regression within masked patches called masked image prediction, which we combined with masked patch token distillation as pretext task to pre-train vision transformers. We show our approach is more accurate and requires fewer fine tuning datasets than other pretext tasks. Unlike prior medical image methods, which typically used image sets arising from disease sites and imaging modalities corresponding to the target tasks, we used 3,643 CT scans (602,708 images) arising from head and neck, lung, and kidney cancers as well as COVID-19 for pre-training and applied it to abdominal organs segmentation from MRI pancreatic cancer patients as well as publicly available 13 different abdominal organs segmentation from CT. Our method showed clear accuracy improvement (average DSC of 0.875 from MRI and 0.878 from CT) with reduced requirement for fine-tuning datasets over commonly used pretext tasks. Extensive comparisons against multiple current SSL methods were done. Code will be made available upon acceptance for publication.
PSIGAN: Joint probabilistic segmentation and image distribution matching for unpaired cross-modality adaptation based MRI segmentation
Jul 18, 2020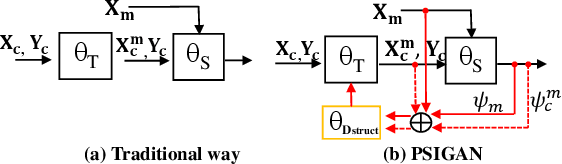
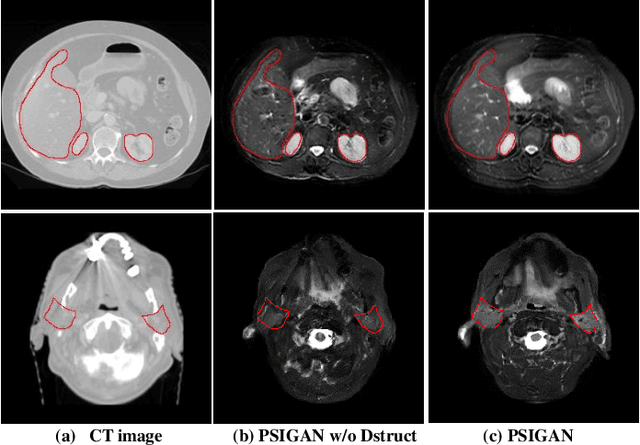
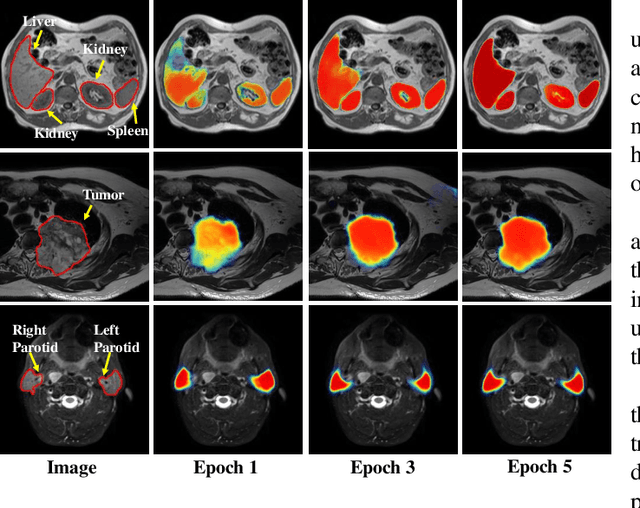
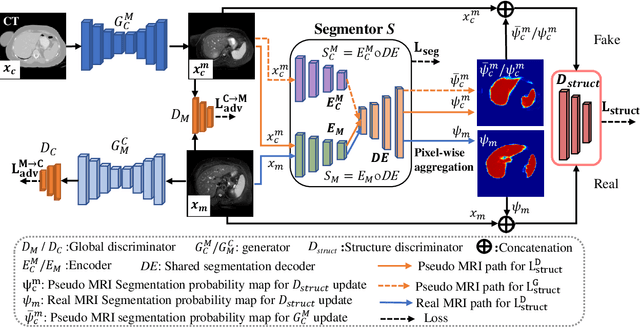
Abstract:We developed a new joint probabilistic segmentation and image distribution matching generative adversarial network (PSIGAN) for unsupervised domain adaptation (UDA) and multi-organ segmentation from magnetic resonance (MRI) images. Our UDA approach models the co-dependency between images and their segmentation as a joint probability distribution using a new structure discriminator. The structure discriminator computes structure of interest focused adversarial loss by combining the generated pseudo MRI with probabilistic segmentations produced by a simultaneously trained segmentation sub-network. The segmentation sub-network is trained using the pseudo MRI produced by the generator sub-network. This leads to a cyclical optimization of both the generator and segmentation sub-networks that are jointly trained as part of an end-to-end network. Extensive experiments and comparisons against multiple state-of-the-art methods were done on four different MRI sequences totalling 257 scans for generating multi-organ and tumor segmentation. The experiments included, (a) 20 T1-weighted (T1w) in-phase mdixon and (b) 20 T2-weighted (T2w) abdominal MRI for segmenting liver, spleen, left and right kidneys, (c) 162 T2-weighted fat suppressed head and neck MRI (T2wFS) for parotid gland segmentation, and (d) 75 T2w MRI for lung tumor segmentation. Our method achieved an overall average DSC of 0.87 on T1w and 0.90 on T2w for the abdominal organs, 0.82 on T2wFS for the parotid glands, and 0.77 on T2w MRI for lung tumors.
* This paper has been accepted by IEEE Transactions on Medical Imaging
Integrating cross-modality hallucinated MRI with CT to aid mediastinal lung tumor segmentation
Sep 10, 2019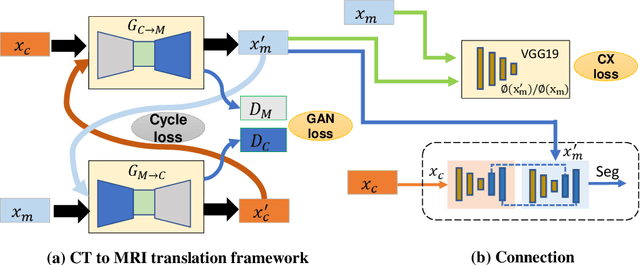
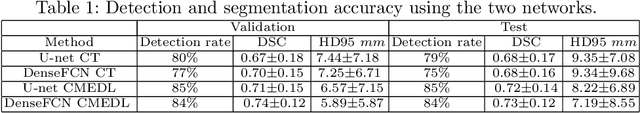
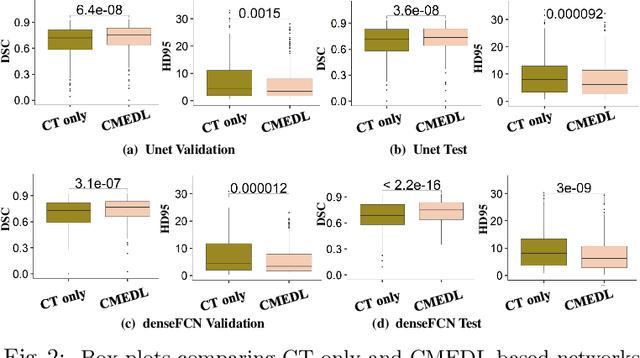
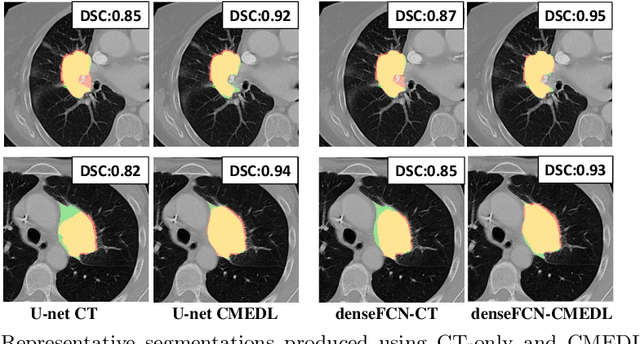
Abstract:Lung tumors, especially those located close to or surrounded by soft tissues like the mediastinum, are difficult to segment due to the low soft tissue contrast on computed tomography images. Magnetic resonance images contain superior soft-tissue contrast information that can be leveraged if both modalities were available for training. Therefore, we developed a cross-modality educed learning approach where MR information that is educed from CT is used to hallucinate MRI and improve CT segmentation. Our approach, called cross-modality educed deep learning segmentation (CMEDL) combines CT and pseudo MR produced from CT by aligning their features to obtain segmentation on CT. Features computed in the last two layers of parallelly trained CT and MR segmentation networks are aligned. We implemented this approach on U-net and dense fully convolutional networks (dense-FCN). Our networks were trained on unrelated cohorts from open-source the Cancer Imaging Archive CT images (N=377), an internal archive T2-weighted MR (N=81), and evaluated using separate validation (N=304) and testing (N=333) CT-delineated tumors. Our approach using both networks were significantly more accurate (U-net $P <0.001$; denseFCN $P <0.001$) than CT-only networks and achieved an accuracy (Dice similarity coefficient) of 0.71$\pm$0.15 (U-net), 0.74$\pm$0.12 (denseFCN) on validation and 0.72$\pm$0.14 (U-net), 0.73$\pm$0.12 (denseFCN) on the testing sets. Our novel approach demonstrated that educing cross-modality information through learned priors enhances CT segmentation performance
Cross-modality (CT-MRI) prior augmented deep learning for robust lung tumor segmentation from small MR datasets
Feb 27, 2019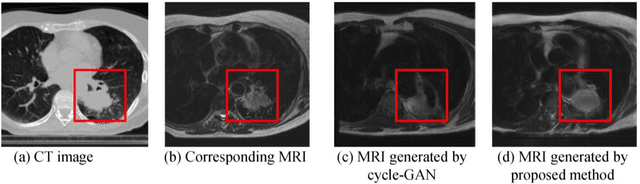
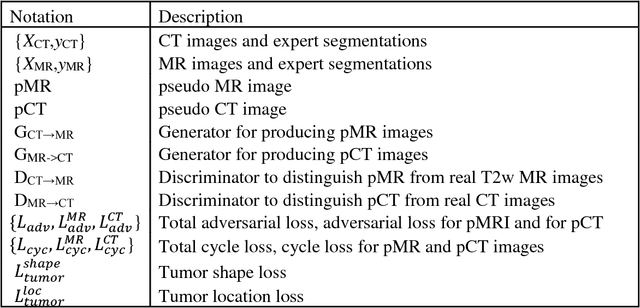
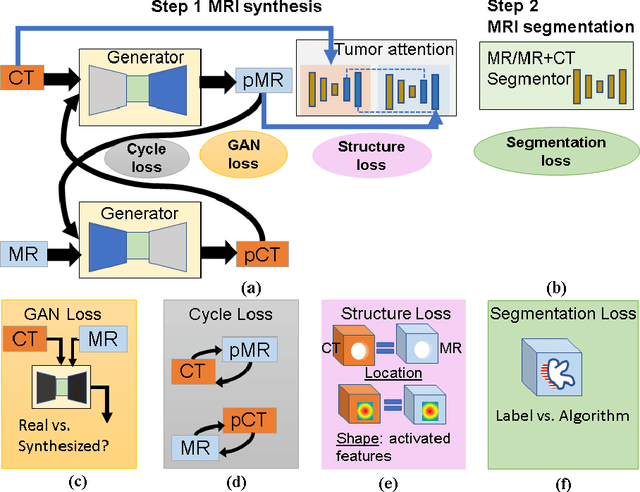
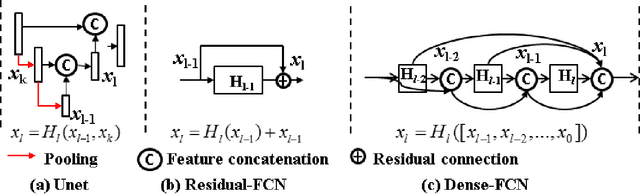
Abstract:Lack of large expert annotated MR datasets makes training deep learning models difficult. Therefore, a cross-modality (MR-CT) deep learning segmentation approach that augments training data using pseudo MR images produced by transforming expert-segmented CT images was developed. Eighty-One T2-weighted MRI scans from 28 patients with non-small cell lung cancers were analyzed. Cross-modality prior encoding the transformation of CT to pseudo MR images resembling T2w MRI was learned as a generative adversarial deep learning model. This model augmented training data arising from 6 expert-segmented T2w MR patient scans with 377 pseudo MRI from non-small cell lung cancer CT patient scans with obtained from the Cancer Imaging Archive. A two-dimensional Unet implemented with batch normalization was trained to segment the tumors from T2w MRI. This method was benchmarked against (a) standard data augmentation and two state-of-the art cross-modality pseudo MR-based augmentation and (b) two segmentation networks. Segmentation accuracy was computed using Dice similarity coefficient (DSC), Hausdroff distance metrics, and volume ratio. The proposed approach produced the lowest statistical variability in the intensity distribution between pseudo and T2w MR images measured as Kullback-Leibler divergence of 0.069. This method produced the highest segmentation accuracy with a DSC of 0.75 and the lowest Hausdroff distance on the test dataset. This approach produced highly similar estimations of tumor growth as an expert (P = 0.37). A novel deep learning MR segmentation was developed that overcomes the limitation of learning robust models from small datasets by leveraging learned cross-modality priors to augment training. The results show the feasibility of the approach and the corresponding improvement over the state-of-the-art methods.
Comparison of Patch-Based Conditional Generative Adversarial Neural Net Models with Emphasis on Model Robustness for Use in Head and Neck Cases for MR-Only planning
Feb 27, 2019



Abstract:A total of twenty paired CT and MR images were used in this study to investigate two conditional generative adversarial networks, Pix2Pix, and Cycle GAN, for generating synthetic CT images for Headand Neck cancer cases. Ten of the patient cases were used for training and included such common artifacts as dental implants; the remaining ten testing cases were used for testing and included a larger range of image features commonly found in clinical head and neck cases. These features included strong metal artifacts from dental implants, one case with a metal implant, and one case with abnormal anatomy. The original CT images were deformably registered to the mDixon FFE MR images to minimize the effects of processing the MR images. The sCT generation accuracy and robustness were evaluated using Mean Absolute Error (MAE) based on the Hounsfield Units (HU) for three regions (whole body, bone, and air within the body), Mean Error (ME) to observe systematic average offset errors in the sCT generation, and dosimetric evaluation of all clinically relevant structures. For the test set the MAE for the Pix2Pix and Cycle GAN models were 92.4 $\pm$ 13.5 HU, and 100.7 $\pm$ 14.6 HU, respectively, for the body region, 166.3 $\pm$ 31.8 HU, and 184 $\pm$ 31.9 HU, respectively, for the bone region, and 183.7 $\pm$ 41.3 HU and 185.4 $\pm$ 37.9 HU for the air regions. The ME for Pix2Pix and Cycle GAN were 21.0 $\pm$ 11.8 HU and 37.5 $\pm$ 14.9 HU, respectively. Absolute Percent Mean/Max Dose Errors were less than 2% for the PTV and all critical structures for both models, and DRRs generated from these models looked qualitatively similar to CT generated DRRs showing these methods are promising for MR-only planning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge