Sean Berry
Optimizing Hard-to-Place Kidney Allocation: A Machine Learning Approach to Center Ranking
Oct 10, 2024Abstract:Kidney transplantation is the preferred treatment for end-stage renal disease, yet the scarcity of donors and inefficiencies in allocation systems create major bottlenecks, resulting in prolonged wait times and alarming mortality rates. Despite their severe scarcity, timely and effective interventions to prevent non-utilization of life-saving organs remain inadequate. Expedited out-of-sequence placement of hard-to-place kidneys to centers with the highest likelihood of utilizing them has been recommended in the literature as an effective strategy to improve placement success. Nevertheless, current attempts towards this practice is non-standardized and heavily rely on the subjective judgment of the decision-makers. This paper proposes a novel data-driven, machine learning-based ranking system for allocating hard-to-place kidneys to centers with a higher likelihood of accepting and successfully transplanting them. Using the national deceased donor kidney offer and transplant datasets, we construct a unique dataset with donor-, center-, and patient-specific features. We propose a data-driven out-of-sequence placement policy that utilizes machine learning models to predict the acceptance probability of a given kidney by a set of transplant centers, ranking them accordingly based on their likelihood of acceptance. Our experiments demonstrate that the proposed policy can reduce the average number of centers considered before placement by fourfold for all kidneys and tenfold for hard-to-place kidneys. This significant reduction indicates that our method can improve the utilization of hard-to-place kidneys and accelerate their acceptance, ultimately reducing patient mortality and the risk of graft failure. Further, we utilize machine learning interpretability tools to provide insights into factors influencing the kidney allocation decisions.
Time Series Clustering for Grouping Products Based on Price and Sales Patterns
Apr 18, 2022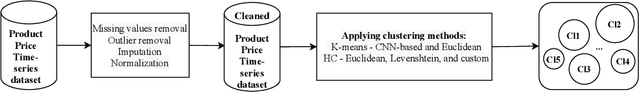
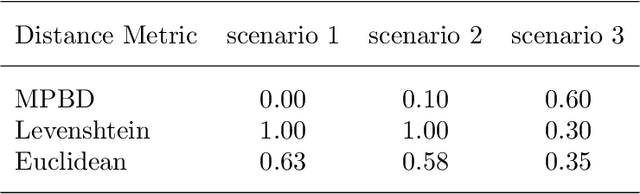
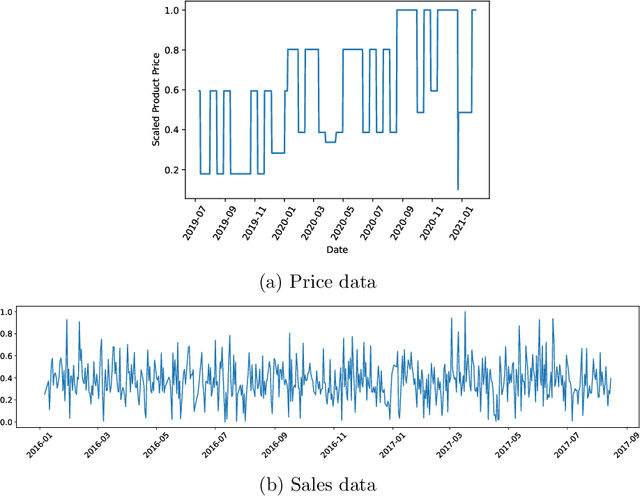
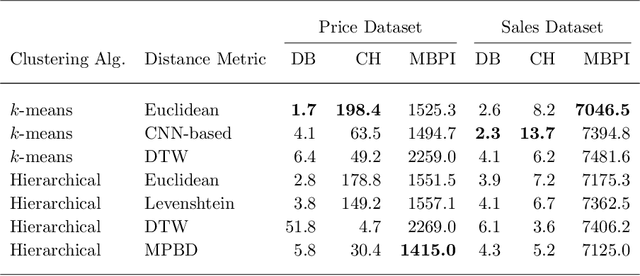
Abstract:Developing technology and changing lifestyles have made online grocery delivery applications an indispensable part of urban life. Since the beginning of the COVID-19 pandemic, the demand for such applications has dramatically increased, creating new competitors that disrupt the market. An increasing level of competition might prompt companies to frequently restructure their marketing and product pricing strategies. Therefore, identifying the change patterns in product prices and sales volumes would provide a competitive advantage for the companies in the marketplace. In this paper, we investigate alternative clustering methodologies to group the products based on the price patterns and sales volumes. We propose a novel distance metric that takes into account how product prices and sales move together rather than calculating the distance using numerical values. We compare our approach with traditional clustering algorithms, which typically rely on generic distance metrics such as Euclidean distance, and image clustering approaches that aim to group data by capturing its visual patterns. We evaluate the performances of different clustering algorithms using our custom evaluation metric as well as Calinski Harabasz and Davies Bouldin indices, which are commonly used internal validity metrics. We conduct our numerical study using a propriety price dataset from an online food and grocery delivery company, and the publicly available Favorita sales dataset. We find that our proposed clustering approach and image clustering both perform well for finding the products with similar price and sales patterns within large datasets.
PSIGAN: Joint probabilistic segmentation and image distribution matching for unpaired cross-modality adaptation based MRI segmentation
Jul 18, 2020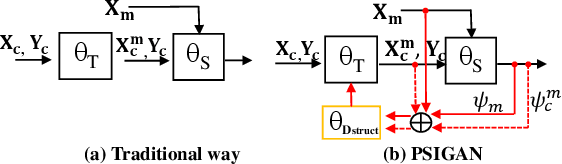
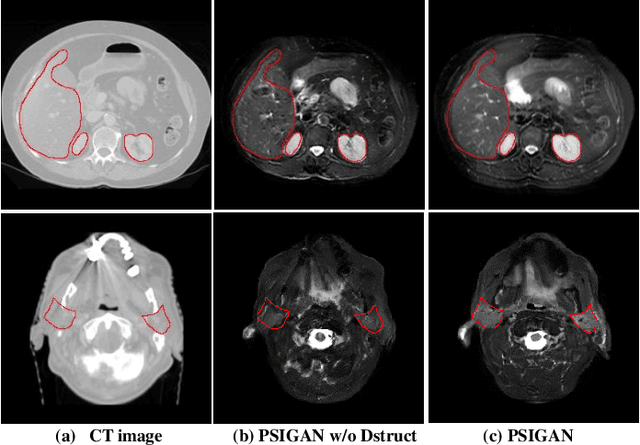
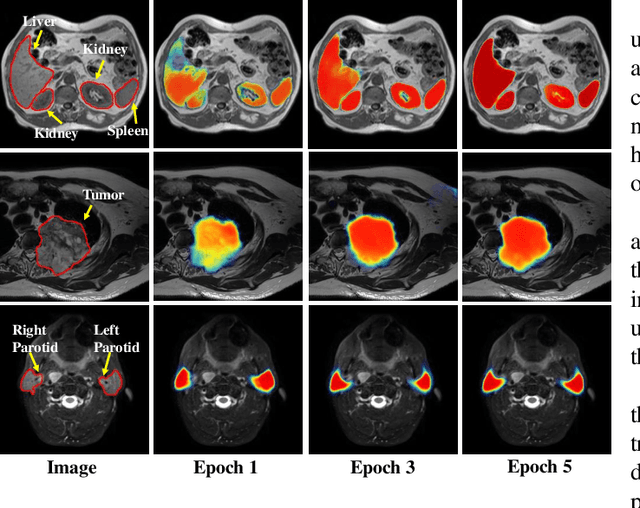
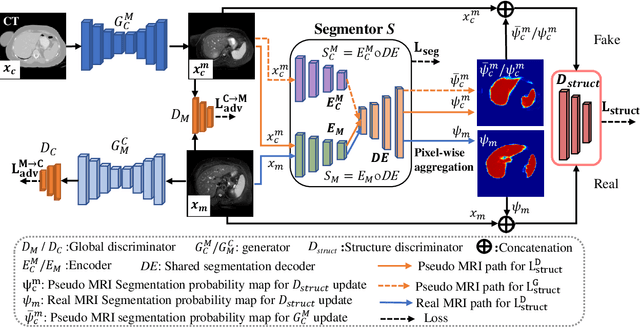
Abstract:We developed a new joint probabilistic segmentation and image distribution matching generative adversarial network (PSIGAN) for unsupervised domain adaptation (UDA) and multi-organ segmentation from magnetic resonance (MRI) images. Our UDA approach models the co-dependency between images and their segmentation as a joint probability distribution using a new structure discriminator. The structure discriminator computes structure of interest focused adversarial loss by combining the generated pseudo MRI with probabilistic segmentations produced by a simultaneously trained segmentation sub-network. The segmentation sub-network is trained using the pseudo MRI produced by the generator sub-network. This leads to a cyclical optimization of both the generator and segmentation sub-networks that are jointly trained as part of an end-to-end network. Extensive experiments and comparisons against multiple state-of-the-art methods were done on four different MRI sequences totalling 257 scans for generating multi-organ and tumor segmentation. The experiments included, (a) 20 T1-weighted (T1w) in-phase mdixon and (b) 20 T2-weighted (T2w) abdominal MRI for segmenting liver, spleen, left and right kidneys, (c) 162 T2-weighted fat suppressed head and neck MRI (T2wFS) for parotid gland segmentation, and (d) 75 T2w MRI for lung tumor segmentation. Our method achieved an overall average DSC of 0.87 on T1w and 0.90 on T2w for the abdominal organs, 0.82 on T2wFS for the parotid glands, and 0.77 on T2w MRI for lung tumors.
* This paper has been accepted by IEEE Transactions on Medical Imaging
Local block-wise self attention for normal organ segmentation
Sep 11, 2019

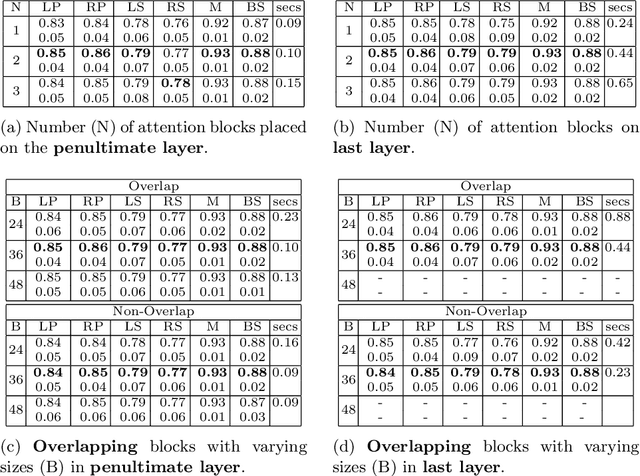

Abstract:We developed a new and computationally simple local block-wise self attention based normal structures segmentation approach applied to head and neck computed tomography (CT) images. Our method uses the insight that normal organs exhibit regularity in their spatial location and inter-relation within images, which can be leveraged to simplify the computations required to aggregate feature information. We accomplish this by using local self attention blocks that pass information between each other to derive the attention map. We show that adding additional attention layers increases the contextual field and captures focused attention from relevant structures. We developed our approach using U-net and compared it against multiple state-of-the-art self attention methods. All models were trained on 48 internal headneck CT scans and tested on 48 CT scans from the external public domain database of computational anatomy dataset. Our method achieved the highest Dice similarity coefficient segmentation accuracy of 0.85$\pm$0.04, 0.86$\pm$0.04 for left and right parotid glands, 0.79$\pm$0.07 and 0.77$\pm$0.05 for left and right submandibular glands, 0.93$\pm$0.01 for mandible and 0.88$\pm$0.02 for the brain stem with the lowest increase of 66.7\% computing time per image and 0.15\% increase in model parameters compared with standard U-net. The best state-of-the-art method called point-wise spatial attention, achieved \textcolor{black}{comparable accuracy but with 516.7\% increase in computing time and 8.14\% increase in parameters compared with standard U-net.} Finally, we performed ablation tests and studied the impact of attention block size, overlap of the attention blocks, additional attention layers, and attention block placement on segmentation performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge