Lei Xing
Reshaping Action Error Distributions for Reliable Vision-Language-Action Models
Feb 04, 2026Abstract:In robotic manipulation, vision-language-action (VLA) models have emerged as a promising paradigm for learning generalizable and scalable robot policies. Most existing VLA frameworks rely on standard supervised objectives, typically cross-entropy for discrete actions and mean squared error (MSE) for continuous action regression, which impose strong pointwise constraints on individual predictions. In this work, we focus on continuous-action VLA models and move beyond conventional MSE-based regression by reshaping action error distributions during training. Drawing on information-theoretic principles, we introduce Minimum Error Entropy (MEE) into modern VLA architectures and propose a trajectory-level MEE objective, together with two weighted variants, combined with MSE for continuous-action VLA training. We evaluate our approaches across standard, few-shot, and noisy settings on multiple representative VLA architectures, using simulation benchmarks such as LIBERO and SimplerEnv as well as real-world robotic manipulation tasks. Experimental results demonstrate consistent improvements in success rates and robustness across these settings. Under imbalanced data regimes, the gains persist within a well-characterized operating range, while incurring negligible additional training cost and no impact on inference efficiency. We further provide theoretical analyses that explain why MEE-based supervision is effective and characterize its practical range. Project Page: https://cognition2actionlab.github.io/VLA-TMEE.github.io/
Latent Reasoning VLA: Latent Thinking and Prediction for Vision-Language-Action Models
Feb 01, 2026Abstract:Vision-Language-Action (VLA) models benefit from chain-of-thought (CoT) reasoning, but existing approaches incur high inference overhead and rely on discrete reasoning representations that mismatch continuous perception and control. We propose Latent Reasoning VLA (\textbf{LaRA-VLA}), a unified VLA framework that internalizes multi-modal CoT reasoning into continuous latent representations for embodied action. LaRA-VLA performs unified reasoning and prediction in latent space, eliminating explicit CoT generation at inference time and enabling efficient, action-oriented control. To realize latent embodied reasoning, we introduce a curriculum-based training paradigm that progressively transitions from explicit textual and visual CoT supervision to latent reasoning, and finally adapts latent reasoning dynamics to condition action generation. We construct two structured CoT datasets and evaluate LaRA-VLA on both simulation benchmarks and long-horizon real-robot manipulation tasks. Experimental results show that LaRA-VLA consistently outperforms state-of-the-art VLA methods while reducing inference latency by up to 90\% compared to explicit CoT-based approaches, demonstrating latent reasoning as an effective and efficient paradigm for real-time embodied control. Project Page: \href{https://loveju1y.github.io/Latent-Reasoning-VLA/}{LaRA-VLA Website}.
Trusted Multi-view Learning for Long-tailed Classification
Nov 12, 2025Abstract:Class imbalance has been extensively studied in single-view scenarios; however, addressing this challenge in multi-view contexts remains an open problem, with even scarcer research focusing on trustworthy solutions. In this paper, we tackle a particularly challenging class imbalance problem in multi-view scenarios: long-tailed classification. We propose TMLC, a Trusted Multi-view Long-tailed Classification framework, which makes contributions on two critical aspects: opinion aggregation and pseudo-data generation. Specifically, inspired by Social Identity Theory, we design a group consensus opinion aggregation mechanism that guides decision making toward the direction favored by the majority of the group. In terms of pseudo-data generation, we introduce a novel distance metric to adapt SMOTE for multi-view scenarios and develop an uncertainty-guided data generation module that produces high-quality pseudo-data, effectively mitigating the adverse effects of class imbalance. Extensive experiments on long-tailed multi-view datasets demonstrate that our model is capable of achieving superior performance. The code is released at https://github.com/cncq-tang/TMLC.
Deep-and-Wide Learning: Enhancing Data-Driven Inference via Synergistic Learning of Inter- and Intra-Data Representations
Jan 28, 2025Abstract:Advancements in deep learning are revolutionizing science and engineering. The immense success of deep learning is largely due to its ability to extract essential high-dimensional (HD) features from input data and make inference decisions based on this information. However, current deep neural network (DNN) models face several challenges, such as the requirements of extensive amounts of data and computational resources. Here, we introduce a new learning scheme, referred to as deep-and-wide learning (DWL), to systematically capture features not only within individual input data (intra-data features) but also across the data (inter-data features). Furthermore, we propose a dual-interactive-channel network (D-Net) to realize the DWL, which leverages our Bayesian formulation of low-dimensional (LD) inter-data feature extraction and its synergistic interaction with the conventional HD representation of the dataset, for substantially enhanced computational efficiency and inference. The proposed technique has been applied to data across various disciplines for both classification and regression tasks. Our results demonstrate that DWL surpasses state-of-the-art DNNs in accuracy by a substantial margin with limited training data and improves the computational efficiency by order(s) of magnitude. The proposed DWL strategy dramatically alters the data-driven learning techniques, including emerging large foundation models, and sheds significant insights into the evolving field of AI.
Generalized Trusted Multi-view Classification Framework with Hierarchical Opinion Aggregation
Nov 06, 2024



Abstract:Recently, multi-view learning has witnessed a considerable interest on the research of trusted decision-making. Previous methods are mainly inspired from an important paper published by Han et al. in 2021, which formulates a Trusted Multi-view Classification (TMC) framework that aggregates evidence from different views based on Dempster's combination rule. All these methods only consider inter-view aggregation, yet lacking exploitation of intra-view information. In this paper, we propose a generalized trusted multi-view classification framework with hierarchical opinion aggregation. This hierarchical framework includes a two-phase aggregation process: the intra-view and inter-view aggregation hierarchies. In the intra aggregation, we assume that each view is comprised of common information shared with other views, as well as its specific information. We then aggregate both the common and specific information. This aggregation phase is useful to eliminate the feature noise inherent to view itself, thereby improving the view quality. In the inter-view aggregation, we design an attention mechanism at the evidence level to facilitate opinion aggregation from different views. To the best of our knowledge, this is one of the pioneering efforts to formulate a hierarchical aggregation framework in the trusted multi-view learning domain. Extensive experiments show that our model outperforms some state-of-art trust-related baselines.
MS-Glance: Non-semantic context vectors and the applications in supervising image reconstruction
Oct 31, 2024Abstract:Non-semantic context information is crucial for visual recognition, as the human visual perception system first uses global statistics to process scenes rapidly before identifying specific objects. However, while semantic information is increasingly incorporated into computer vision tasks such as image reconstruction, non-semantic information, such as global spatial structures, is often overlooked. To bridge the gap, we propose a biologically informed non-semantic context descriptor, \textbf{MS-Glance}, along with the Glance Index Measure for comparing two images. A Global Glance vector is formulated by randomly retrieving pixels based on a perception-driven rule from an image to form a vector representing non-semantic global context, while a local Glance vector is a flattened local image window, mimicking a zoom-in observation. The Glance Index is defined as the inner product of two standardized sets of Glance vectors. We evaluate the effectiveness of incorporating Glance supervision in two reconstruction tasks: image fitting with implicit neural representation (INR) and undersampled MRI reconstruction. Extensive experimental results show that MS-Glance outperforms existing image restoration losses across both natural and medical images. The code is available at \url{https://github.com/Z7Gao/MSGlance}.
ST-NeRP: Spatial-Temporal Neural Representation Learning with Prior Embedding for Patient-specific Imaging Study
Oct 25, 2024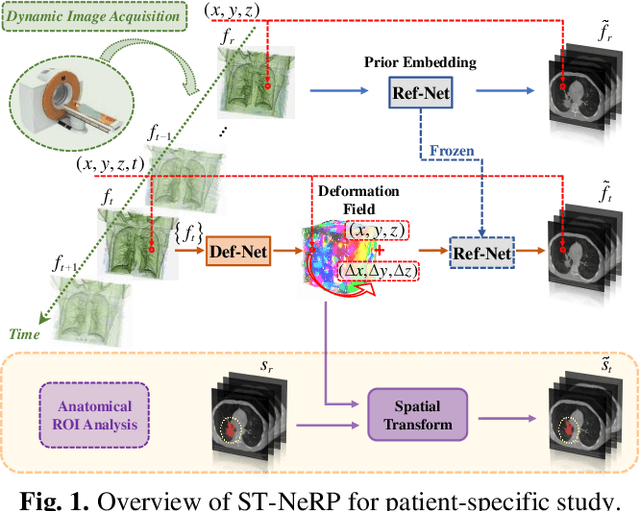
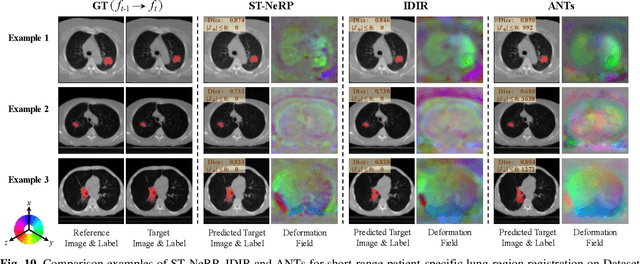
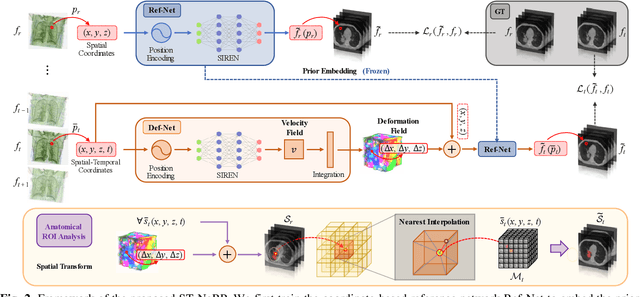
Abstract:During and after a course of therapy, imaging is routinely used to monitor the disease progression and assess the treatment responses. Despite of its significance, reliably capturing and predicting the spatial-temporal anatomic changes from a sequence of patient-specific image series presents a considerable challenge. Thus, the development of a computational framework becomes highly desirable for a multitude of practical applications. In this context, we propose a strategy of Spatial-Temporal Neural Representation learning with Prior embedding (ST-NeRP) for patient-specific imaging study. Our strategy involves leveraging an Implicit Neural Representation (INR) network to encode the image at the reference time point into a prior embedding. Subsequently, a spatial-temporally continuous deformation function is learned through another INR network. This network is trained using the whole patient-specific image sequence, enabling the prediction of deformation fields at various target time points. The efficacy of the ST-NeRP model is demonstrated through its application to diverse sequential image series, including 4D CT and longitudinal CT datasets within thoracic and abdominal imaging. The proposed ST-NeRP model exhibits substantial potential in enabling the monitoring of anatomical changes within a patient throughout the therapeutic journey.
Discovering distinctive elements of biomedical datasets for high-performance exploration
Oct 07, 2024



Abstract:The human brain represents an object by small elements and distinguishes two objects based on the difference in elements. Discovering the distinctive elements of high-dimensional datasets is therefore critical in numerous perception-driven biomedical and clinical studies. However, currently there is no available method for reliable extraction of distinctive elements of high-dimensional biomedical and clinical datasets. Here we present an unsupervised deep learning technique namely distinctive element analysis (DEA), which extracts the distinctive data elements using high-dimensional correlative information of the datasets. DEA at first computes a large number of distinctive parts of the data, then filters and condenses the parts into DEA elements by employing a unique kernel-driven triple-optimization network. DEA has been found to improve the accuracy by up to 45% in comparison to the traditional techniques in applications such as disease detection from medical images, gene ranking and cell recognition from single cell RNA sequence (scRNA-seq) datasets. Moreover, DEA allows user-guided manipulation of the intermediate calculation process and thus offers intermediate results with better interpretability.
Multi-sensor Learning Enables Information Transfer across Different Sensory Data and Augments Multi-modality Imaging
Sep 28, 2024Abstract:Multi-modality imaging is widely used in clinical practice and biomedical research to gain a comprehensive understanding of an imaging subject. Currently, multi-modality imaging is accomplished by post hoc fusion of independently reconstructed images under the guidance of mutual information or spatially registered hardware, which limits the accuracy and utility of multi-modality imaging. Here, we investigate a data-driven multi-modality imaging (DMI) strategy for synergetic imaging of CT and MRI. We reveal two distinct types of features in multi-modality imaging, namely intra- and inter-modality features, and present a multi-sensor learning (MSL) framework to utilize the crossover inter-modality features for augmented multi-modality imaging. The MSL imaging approach breaks down the boundaries of traditional imaging modalities and allows for optimal hybridization of CT and MRI, which maximizes the use of sensory data. We showcase the effectiveness of our DMI strategy through synergetic CT-MRI brain imaging. The principle of DMI is quite general and holds enormous potential for various DMI applications across disciplines.
The Application of Machine Learning in Tidal Evolution Simulation of Star-Planet Systems
Aug 29, 2024



Abstract:With the release of a large amount of astronomical data, an increasing number of close-in hot Jupiters have been discovered. Calculating their evolutionary curves using star-planet interaction models presents a challenge. To expedite the generation of evolutionary curves for these close-in hot Jupiter systems, we utilized tidal interaction models established on MESA to create 15,745 samples of star-planet systems and 7,500 samples of stars. Additionally, we employed a neural network (Multi-Layer Perceptron - MLP) to predict the evolutionary curves of the systems, including stellar effective temperature, radius, stellar rotation period, and planetary orbital period. The median relative errors of the predicted evolutionary curves were found to be 0.15%, 0.43%, 2.61%, and 0.57%, respectively. Furthermore, the speed at which we generate evolutionary curves exceeds that of model-generated curves by more than four orders of magnitude. We also extracted features of planetary migration states and utilized lightGBM to classify the samples into 6 categories for prediction. We found that by combining three types that undergo long-term double synchronization into one label, the classifier effectively recognized these features. Apart from systems experiencing long-term double synchronization, the median relative errors of the predicted evolutionary curves were all below 4%. Our work provides an efficient method to save significant computational resources and time with minimal loss in accuracy. This research also lays the foundation for analyzing the evolutionary characteristics of systems under different migration states, aiding in the understanding of the underlying physical mechanisms of such systems. Finally, to a large extent, our approach could replace the calculations of theoretical models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge