Michael Gensheimer
Large Language Model-Augmented Auto-Delineation of Treatment Target Volume in Radiation Therapy
Jul 10, 2024Abstract:Radiation therapy (RT) is one of the most effective treatments for cancer, and its success relies on the accurate delineation of targets. However, target delineation is a comprehensive medical decision that currently relies purely on manual processes by human experts. Manual delineation is time-consuming, laborious, and subject to interobserver variations. Although the advancements in artificial intelligence (AI) techniques have significantly enhanced the auto-contouring of normal tissues, accurate delineation of RT target volumes remains a challenge. In this study, we propose a visual language model-based RT target volume auto-delineation network termed Radformer. The Radformer utilizes a hierarichal vision transformer as the backbone and incorporates large language models to extract text-rich features from clinical data. We introduce a visual language attention module (VLAM) for integrating visual and linguistic features for language-aware visual encoding (LAVE). The Radformer has been evaluated on a dataset comprising 2985 patients with head-and-neck cancer who underwent RT. Metrics, including the Dice similarity coefficient (DSC), intersection over union (IOU), and 95th percentile Hausdorff distance (HD95), were used to evaluate the performance of the model quantitatively. Our results demonstrate that the Radformer has superior segmentation performance compared to other state-of-the-art models, validating its potential for adoption in RT practice.
Automated radiotherapy treatment planning guided by GPT-4Vision
Jun 21, 2024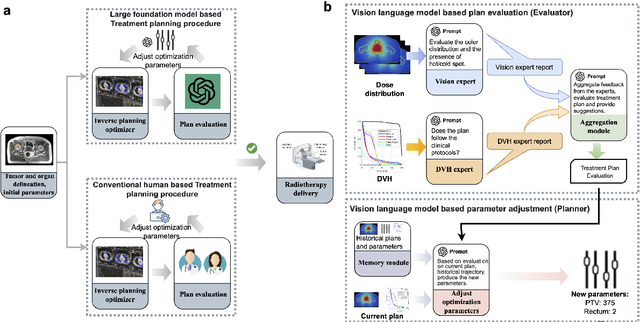
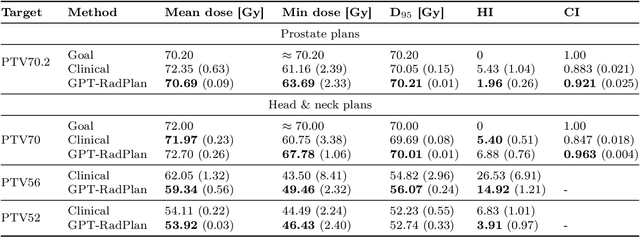
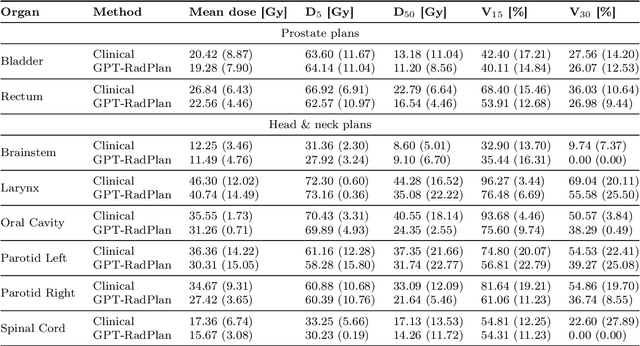
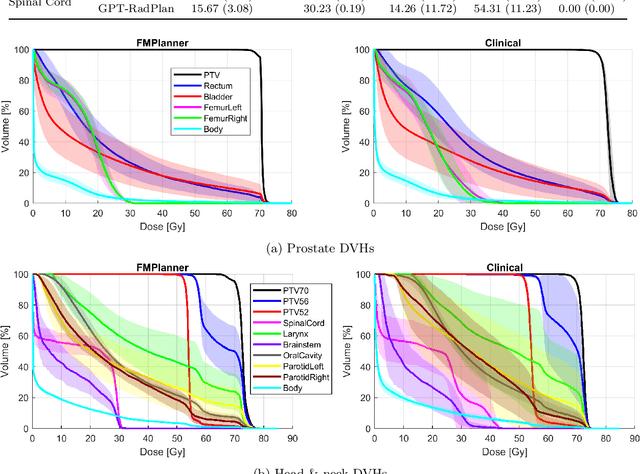
Abstract:Radiotherapy treatment planning is a time-consuming and potentially subjective process that requires the iterative adjustment of model parameters to balance multiple conflicting objectives. Recent advancements in large foundation models offer promising avenues for addressing the challenges in planning and clinical decision-making. This study introduces GPT-RadPlan, a fully automated treatment planning framework that harnesses prior radiation oncology knowledge encoded in multi-modal large language models, such as GPT-4Vision (GPT-4V) from OpenAI. GPT-RadPlan is made aware of planning protocols as context and acts as an expert human planner, capable of guiding a treatment planning process. Via in-context learning, we incorporate clinical protocols for various disease sites as prompts to enable GPT-4V to acquire treatment planning domain knowledge. The resulting GPT-RadPlan agent is integrated into our in-house inverse treatment planning system through an API. The efficacy of the automated planning system is showcased using multiple prostate and head & neck cancer cases, where we compared GPT-RadPlan results to clinical plans. In all cases, GPT-RadPlan either outperformed or matched the clinical plans, demonstrating superior target coverage and organ-at-risk sparing. Consistently satisfying the dosimetric objectives in the clinical protocol, GPT-RadPlan represents the first multimodal large language model agent that mimics the behaviors of human planners in radiation oncology clinics, achieving remarkable results in automating the treatment planning process without the need for additional training.
Atlas Based Segmentations via Semi-Supervised Diffeomorphic Registrations
Nov 23, 2019



Abstract:Purpose: Segmentation of organs-at-risk (OARs) is a bottleneck in current radiation oncology pipelines and is often time consuming and labor intensive. In this paper, we propose an atlas-based semi-supervised registration algorithm to generate accurate segmentations of OARs for which there are ground truth contours and rough segmentations of all other OARs in the atlas. To the best of our knowledge, this is the first study to use learning-based registration methods for the segmentation of head and neck patients and demonstrate its utility in clinical applications. Methods: Our algorithm cascades rigid and deformable deformation blocks, and takes on an atlas image (M), set of atlas-space segmentations (S_A), and a patient image (F) as inputs, while outputting patient-space segmentations of all OARs defined on the atlas. We train our model on 475 CT images taken from public archives and Stanford RadOnc Clinic (SROC), validate on 5 CT images from SROC, and test our model on 20 CT images from SROC. Results: Our method outperforms current state of the art learning-based registration algorithms and achieves an overall dice score of 0.789 on our test set. Moreover, our method yields a performance comparable to manual segmentation and supervised segmentation, while solving a much more complex registration problem. Whereas supervised segmentation methods only automate the segmentation process for a select few number of OARs, we demonstrate that our methods can achieve similar performance for OARs of interest, while also providing segmentations for every other OAR on the provided atlas. Conclusions: Our proposed algorithm has significant clinical applications and could help reduce the bottleneck for segmentation of head and neck OARs. Further, our results demonstrate that semi-supervised diffeomorphic registration can be accurately applied to both registration and segmentation problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge