Xiaokun Liang
Data-Efficient Meningioma Segmentation via Implicit Spatiotemporal Mixing and Sim2Real Semantic Injection
Jan 19, 2026Abstract:The performance of medical image segmentation is increasingly defined by the efficiency of data utilization rather than merely the volume of raw data. Accurate segmentation, particularly for complex pathologies like meningiomas, demands that models fully exploit the latent information within limited high-quality annotations. To maximize the value of existing datasets, we propose a novel dual-augmentation framework that synergistically integrates spatial manifold expansion and semantic object injection. Specifically, we leverage Implicit Neural Representations (INR) to model continuous velocity fields. Unlike previous methods, we perform linear mixing on the integrated deformation fields, enabling the efficient generation of anatomically plausible variations by interpolating within the deformation space. This approach allows for the extensive exploration of structural diversity from a small set of anchors. Furthermore, we introduce a Sim2Real lesion injection module. This module constructs a high-fidelity simulation domain by transplanting lesion textures into healthy anatomical backgrounds, effectively bridging the gap between synthetic augmentation and real-world pathology. Comprehensive experiments on a hybrid dataset demonstrate that our framework significantly enhances the data efficiency and robustness of state-of-the-art models, including nnU-Net and U-Mamba, offering a potent strategy for high-performance medical image analysis with limited annotation budgets.
Calibration and Uncertainty for multiRater Volume Assessment in multiorgan Segmentation (CURVAS) challenge results
May 13, 2025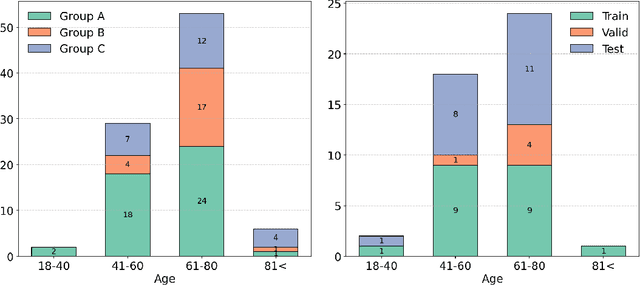
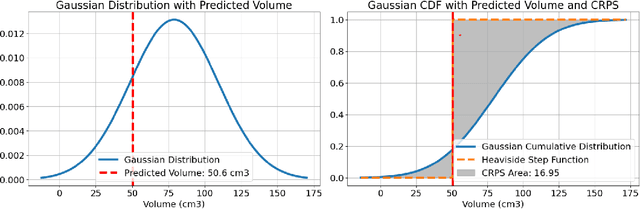

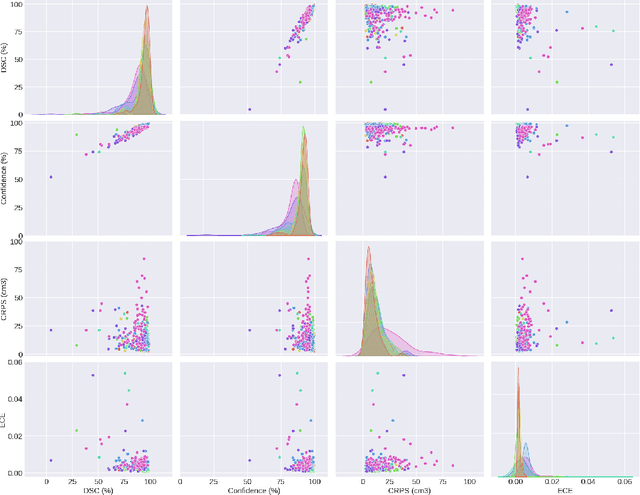
Abstract:Deep learning (DL) has become the dominant approach for medical image segmentation, yet ensuring the reliability and clinical applicability of these models requires addressing key challenges such as annotation variability, calibration, and uncertainty estimation. This is why we created the Calibration and Uncertainty for multiRater Volume Assessment in multiorgan Segmentation (CURVAS), which highlights the critical role of multiple annotators in establishing a more comprehensive ground truth, emphasizing that segmentation is inherently subjective and that leveraging inter-annotator variability is essential for robust model evaluation. Seven teams participated in the challenge, submitting a variety of DL models evaluated using metrics such as Dice Similarity Coefficient (DSC), Expected Calibration Error (ECE), and Continuous Ranked Probability Score (CRPS). By incorporating consensus and dissensus ground truth, we assess how DL models handle uncertainty and whether their confidence estimates align with true segmentation performance. Our findings reinforce the importance of well-calibrated models, as better calibration is strongly correlated with the quality of the results. Furthermore, we demonstrate that segmentation models trained on diverse datasets and enriched with pre-trained knowledge exhibit greater robustness, particularly in cases deviating from standard anatomical structures. Notably, the best-performing models achieved high DSC and well-calibrated uncertainty estimates. This work underscores the need for multi-annotator ground truth, thorough calibration assessments, and uncertainty-aware evaluations to develop trustworthy and clinically reliable DL-based medical image segmentation models.
Benchmark of Segmentation Techniques for Pelvic Fracture in CT and X-ray: Summary of the PENGWIN 2024 Challenge
Apr 03, 2025Abstract:The segmentation of pelvic fracture fragments in CT and X-ray images is crucial for trauma diagnosis, surgical planning, and intraoperative guidance. However, accurately and efficiently delineating the bone fragments remains a significant challenge due to complex anatomy and imaging limitations. The PENGWIN challenge, organized as a MICCAI 2024 satellite event, aimed to advance automated fracture segmentation by benchmarking state-of-the-art algorithms on these complex tasks. A diverse dataset of 150 CT scans was collected from multiple clinical centers, and a large set of simulated X-ray images was generated using the DeepDRR method. Final submissions from 16 teams worldwide were evaluated under a rigorous multi-metric testing scheme. The top-performing CT algorithm achieved an average fragment-wise intersection over union (IoU) of 0.930, demonstrating satisfactory accuracy. However, in the X-ray task, the best algorithm attained an IoU of 0.774, highlighting the greater challenges posed by overlapping anatomical structures. Beyond the quantitative evaluation, the challenge revealed methodological diversity in algorithm design. Variations in instance representation, such as primary-secondary classification versus boundary-core separation, led to differing segmentation strategies. Despite promising results, the challenge also exposed inherent uncertainties in fragment definition, particularly in cases of incomplete fractures. These findings suggest that interactive segmentation approaches, integrating human decision-making with task-relevant information, may be essential for improving model reliability and clinical applicability.
Multi-Class Segmentation of Aortic Branches and Zones in Computed Tomography Angiography: The AortaSeg24 Challenge
Feb 07, 2025



Abstract:Multi-class segmentation of the aorta in computed tomography angiography (CTA) scans is essential for diagnosing and planning complex endovascular treatments for patients with aortic dissections. However, existing methods reduce aortic segmentation to a binary problem, limiting their ability to measure diameters across different branches and zones. Furthermore, no open-source dataset is currently available to support the development of multi-class aortic segmentation methods. To address this gap, we organized the AortaSeg24 MICCAI Challenge, introducing the first dataset of 100 CTA volumes annotated for 23 clinically relevant aortic branches and zones. This dataset was designed to facilitate both model development and validation. The challenge attracted 121 teams worldwide, with participants leveraging state-of-the-art frameworks such as nnU-Net and exploring novel techniques, including cascaded models, data augmentation strategies, and custom loss functions. We evaluated the submitted algorithms using the Dice Similarity Coefficient (DSC) and Normalized Surface Distance (NSD), highlighting the approaches adopted by the top five performing teams. This paper presents the challenge design, dataset details, evaluation metrics, and an in-depth analysis of the top-performing algorithms. The annotated dataset, evaluation code, and implementations of the leading methods are publicly available to support further research. All resources can be accessed at https://aortaseg24.grand-challenge.org.
Three-Dimensional Medical Image Fusion with Deformable Cross-Attention
Oct 10, 2023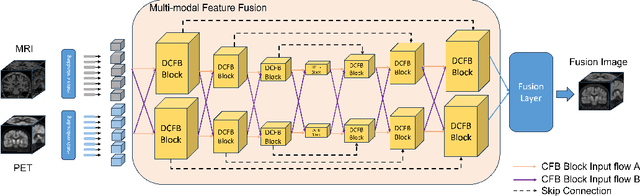
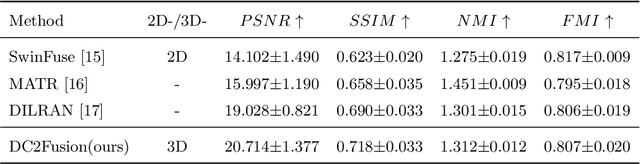
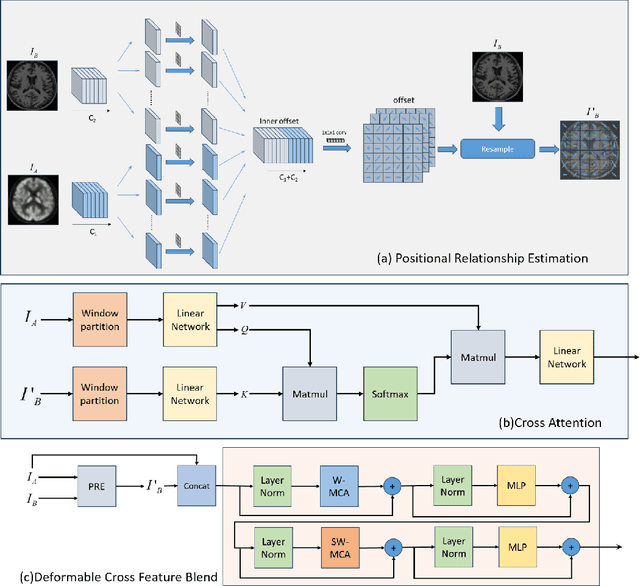
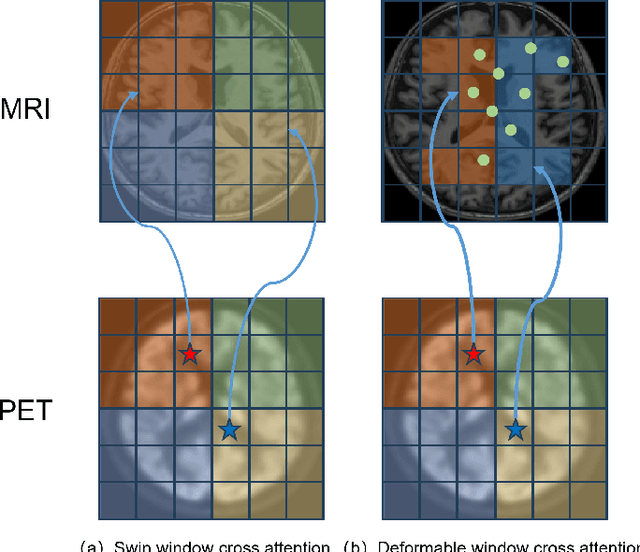
Abstract:Multimodal medical image fusion plays an instrumental role in several areas of medical image processing, particularly in disease recognition and tumor detection. Traditional fusion methods tend to process each modality independently before combining the features and reconstructing the fusion image. However, this approach often neglects the fundamental commonalities and disparities between multimodal information. Furthermore, the prevailing methodologies are largely confined to fusing two-dimensional (2D) medical image slices, leading to a lack of contextual supervision in the fusion images and subsequently, a decreased information yield for physicians relative to three-dimensional (3D) images. In this study, we introduce an innovative unsupervised feature mutual learning fusion network designed to rectify these limitations. Our approach incorporates a Deformable Cross Feature Blend (DCFB) module that facilitates the dual modalities in discerning their respective similarities and differences. We have applied our model to the fusion of 3D MRI and PET images obtained from 660 patients in the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset. Through the application of the DCFB module, our network generates high-quality MRI-PET fusion images. Experimental results demonstrate that our method surpasses traditional 2D image fusion methods in performance metrics such as Peak Signal to Noise Ratio (PSNR) and Structural Similarity Index Measure (SSIM). Importantly, the capacity of our method to fuse 3D images enhances the information available to physicians and researchers, thus marking a significant step forward in the field. The code will soon be available online.
QUIZ: An Arbitrary Volumetric Point Matching Method for Medical Image Registration
Sep 30, 2023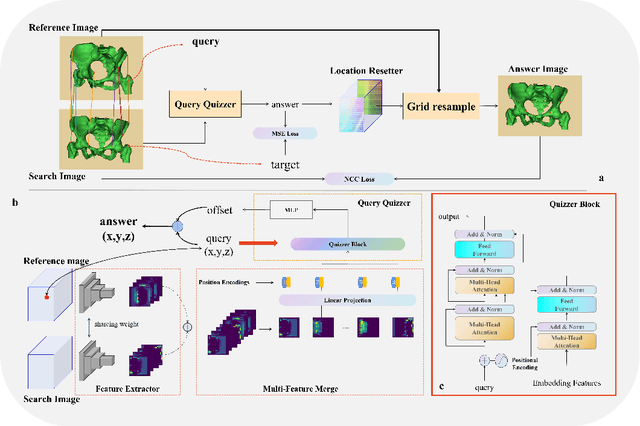
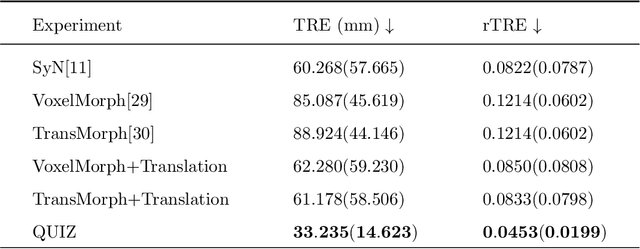
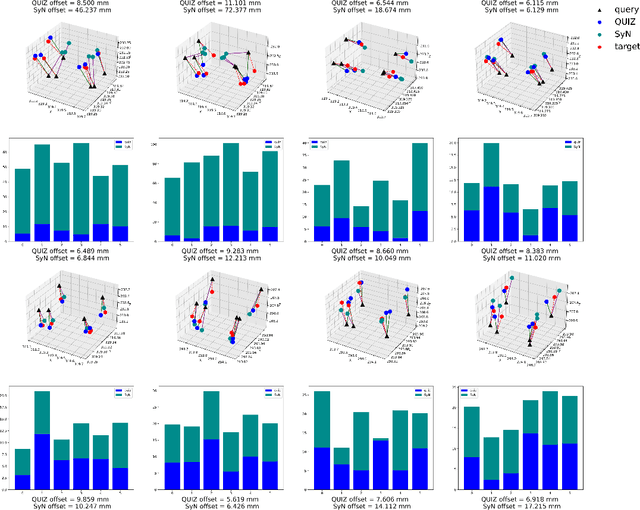
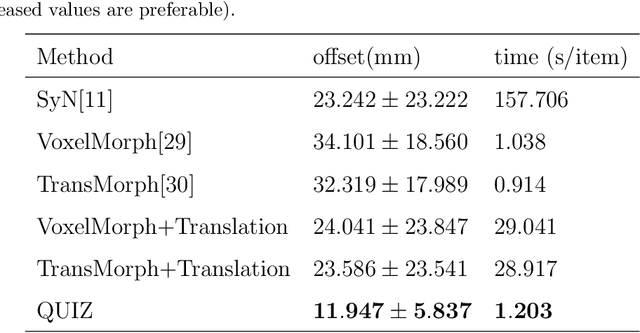
Abstract:Rigid pre-registration involving local-global matching or other large deformation scenarios is crucial. Current popular methods rely on unsupervised learning based on grayscale similarity, but under circumstances where different poses lead to varying tissue structures, or where image quality is poor, these methods tend to exhibit instability and inaccuracies. In this study, we propose a novel method for medical image registration based on arbitrary voxel point of interest matching, called query point quizzer (QUIZ). QUIZ focuses on the correspondence between local-global matching points, specifically employing CNN for feature extraction and utilizing the Transformer architecture for global point matching queries, followed by applying average displacement for local image rigid transformation. We have validated this approach on a large deformation dataset of cervical cancer patients, with results indicating substantially smaller deviations compared to state-of-the-art methods. Remarkably, even for cross-modality subjects, it achieves results surpassing the current state-of-the-art.
Unsupervised CT Metal Artifact Reduction by Plugging Diffusion Priors in Dual Domains
Aug 31, 2023



Abstract:During the process of computed tomography (CT), metallic implants often cause disruptive artifacts in the reconstructed images, impeding accurate diagnosis. Several supervised deep learning-based approaches have been proposed for reducing metal artifacts (MAR). However, these methods heavily rely on training with simulated data, as obtaining paired metal artifact CT and clean CT data in clinical settings is challenging. This limitation can lead to decreased performance when applying these methods in clinical practice. Existing unsupervised MAR methods, whether based on learning or not, typically operate within a single domain, either in the image domain or the sinogram domain. In this paper, we propose an unsupervised MAR method based on the diffusion model, a generative model with a high capacity to represent data distributions. Specifically, we first train a diffusion model using CT images without metal artifacts. Subsequently, we iteratively utilize the priors embedded within the pre-trained diffusion model in both the sinogram and image domains to restore the degraded portions caused by metal artifacts. This dual-domain processing empowers our approach to outperform existing unsupervised MAR methods, including another MAR method based on the diffusion model, which we have qualitatively and quantitatively validated using synthetic datasets. Moreover, our method demonstrates superior visual results compared to both supervised and unsupervised methods on clinical datasets.
INR-LDDMM: Fluid-based Medical Image Registration Integrating Implicit Neural Representation and Large Deformation Diffeomorphic Metric Mapping
Aug 21, 2023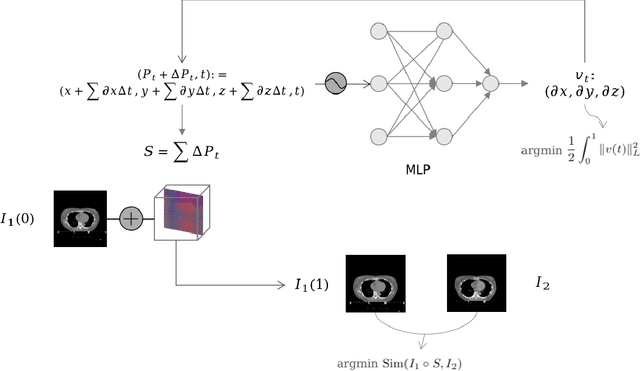
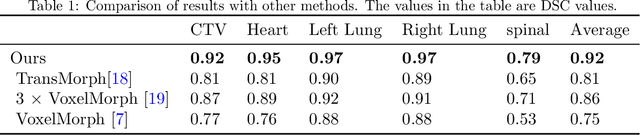
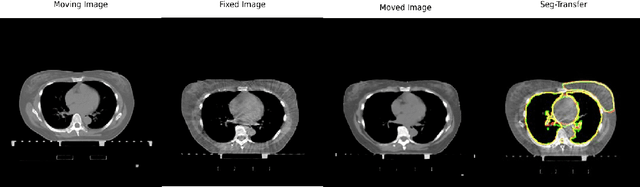
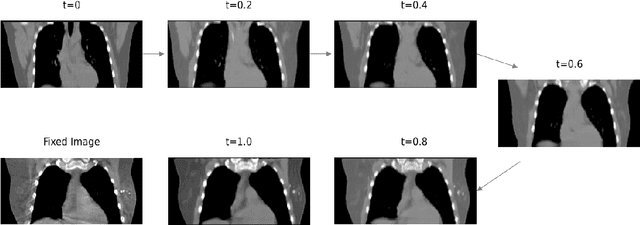
Abstract:We propose a flow-based registration framework of medical images based on implicit neural representation. By integrating implicit neural representation and Large Deformable Diffeomorphic Metric Mapping (LDDMM), we employ a Multilayer Perceptron (MLP) as a velocity generator while optimizing velocity and image similarity. Moreover, we adopt a coarse-to-fine approach to address the challenge of deformable-based registration methods dropping into local optimal solutions, thus aiding the management of significant deformations in medical image registration. Our algorithm has been validated on a paired CT-CBCT dataset of 50 patients,taking the dice coefficient of transferred annotations as an evaluation metric. Compared to existing methods, our approach achieves the state-of-the-art performance.
XTransCT: Ultra-Fast Volumetric CT Reconstruction using Two Orthogonal X-Ray Projections via a Transformer Network
May 31, 2023Abstract:Computed tomography (CT) scans offer a detailed, three-dimensional representation of patients' internal organs. However, conventional CT reconstruction techniques necessitate acquiring hundreds or thousands of x-ray projections through a complete rotational scan of the body, making navigation or positioning during surgery infeasible. In image-guided radiation therapy, a method that reconstructs ultra-sparse X-ray projections into CT images, we can exploit the substantially reduced radiation dose and minimize equipment burden for localization and navigation. In this study, we introduce a novel Transformer architecture, termed XTransCT, devised to facilitate real-time reconstruction of CT images from two-dimensional X-ray images. We assess our approach regarding image quality and structural reliability using a dataset of fifty patients, supplied by a hospital, as well as the larger public dataset LIDC-IDRI, which encompasses thousands of patients. Additionally, we validated our algorithm's generalizability on the LNDb dataset. Our findings indicate that our algorithm surpasses other methods in image quality, structural precision, and generalizability. Moreover, in comparison to previous 3D convolution-based approaches, we note a substantial speed increase of approximately 300 $\%$, achieving 44 ms per 3D image reconstruction. To guarantee the replicability of our results, we have made our code publicly available.
A Diffusion Probabilistic Prior for Low-Dose CT Image Denoising
May 25, 2023Abstract:Low-dose computed tomography (CT) image denoising is crucial in medical image computing. Recent years have been remarkable improvement in deep learning-based methods for this task. However, training deep denoising neural networks requires low-dose and normal-dose CT image pairs, which are difficult to obtain in the clinic settings. To address this challenge, we propose a novel fully unsupervised method for low-dose CT image denoising, which is based on denoising diffusion probabilistic model -- a powerful generative model. First, we train an unconditional denoising diffusion probabilistic model capable of generating high-quality normal-dose CT images from random noise. Subsequently, the probabilistic priors of the pre-trained diffusion model are incorporated into a Maximum A Posteriori (MAP) estimation framework for iteratively solving the image denoising problem. Our method ensures the diffusion model produces high-quality normal-dose CT images while keeping the image content consistent with the input low-dose CT images. We evaluate our method on a widely used low-dose CT image denoising benchmark, and it outperforms several supervised low-dose CT image denoising methods in terms of both quantitative and visual performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge