Haoyang Liu
PPI-SVRG: Unifying Prediction-Powered Inference and Variance Reduction for Semi-Supervised Optimization
Jan 29, 2026Abstract:We study semi-supervised stochastic optimization when labeled data is scarce but predictions from pre-trained models are available. PPI and SVRG both reduce variance through control variates -- PPI uses predictions, SVRG uses reference gradients. We show they are mathematically equivalent and develop PPI-SVRG, which combines both. Our convergence bound decomposes into the standard SVRG rate plus an error floor from prediction uncertainty. The rate depends only on loss geometry; predictions affect only the neighborhood size. When predictions are perfect, we recover SVRG exactly. When predictions degrade, convergence remains stable but reaches a larger neighborhood. Experiments confirm the theory: PPI-SVRG reduces MSE by 43--52\% under label scarcity on mean estimation benchmarks and improves test accuracy by 2.7--2.9 percentage points on MNIST with only 10\% labeled data.
CoCo-MILP: Inter-Variable Contrastive and Intra-Constraint Competitive MILP Solution Prediction
Nov 12, 2025Abstract:Mixed-Integer Linear Programming (MILP) is a cornerstone of combinatorial optimization, yet solving large-scale instances remains a significant computational challenge. Recently, Graph Neural Networks (GNNs) have shown promise in accelerating MILP solvers by predicting high-quality solutions. However, we identify that existing methods misalign with the intrinsic structure of MILP problems at two levels. At the leaning objective level, the Binary Cross-Entropy (BCE) loss treats variables independently, neglecting their relative priority and yielding plausible logits. At the model architecture level, standard GNN message passing inherently smooths the representations across variables, missing the natural competitive relationships within constraints. To address these challenges, we propose CoCo-MILP, which explicitly models inter-variable Contrast and intra-constraint Competition for advanced MILP solution prediction. At the objective level, CoCo-MILP introduces the Inter-Variable Contrastive Loss (VCL), which explicitly maximizes the embedding margin between variables assigned one versus zero. At the architectural level, we design an Intra-Constraint Competitive GNN layer that, instead of homogenizing features, learns to differentiate representations of competing variables within a constraint, capturing their exclusionary nature. Experimental results on standard benchmarks demonstrate that CoCo-MILP significantly outperforms existing learning-based approaches, reducing the solution gap by up to 68.12% compared to traditional solvers. Our code is available at https://github.com/happypu326/CoCo-MILP.
GenoMAS: A Multi-Agent Framework for Scientific Discovery via Code-Driven Gene Expression Analysis
Jul 28, 2025Abstract:Gene expression analysis holds the key to many biomedical discoveries, yet extracting insights from raw transcriptomic data remains formidable due to the complexity of multiple large, semi-structured files and the need for extensive domain expertise. Current automation approaches are often limited by either inflexible workflows that break down in edge cases or by fully autonomous agents that lack the necessary precision for rigorous scientific inquiry. GenoMAS charts a different course by presenting a team of LLM-based scientists that integrates the reliability of structured workflows with the adaptability of autonomous agents. GenoMAS orchestrates six specialized LLM agents through typed message-passing protocols, each contributing complementary strengths to a shared analytic canvas. At the heart of GenoMAS lies a guided-planning framework: programming agents unfold high-level task guidelines into Action Units and, at each juncture, elect to advance, revise, bypass, or backtrack, thereby maintaining logical coherence while bending gracefully to the idiosyncrasies of genomic data. On the GenoTEX benchmark, GenoMAS reaches a Composite Similarity Correlation of 89.13% for data preprocessing and an F$_1$ of 60.48% for gene identification, surpassing the best prior art by 10.61% and 16.85% respectively. Beyond metrics, GenoMAS surfaces biologically plausible gene-phenotype associations corroborated by the literature, all while adjusting for latent confounders. Code is available at https://github.com/Liu-Hy/GenoMAS.
Apollo-MILP: An Alternating Prediction-Correction Neural Solving Framework for Mixed-Integer Linear Programming
Mar 03, 2025Abstract:Leveraging machine learning (ML) to predict an initial solution for mixed-integer linear programming (MILP) has gained considerable popularity in recent years. These methods predict a solution and fix a subset of variables to reduce the problem dimension. Then, they solve the reduced problem to obtain the final solutions. However, directly fixing variable values can lead to low-quality solutions or even infeasible reduced problems if the predicted solution is not accurate enough. To address this challenge, we propose an Alternating prediction-correction neural solving framework (Apollo-MILP) that can identify and select accurate and reliable predicted values to fix. In each iteration, Apollo-MILP conducts a prediction step for the unfixed variables, followed by a correction step to obtain an improved solution (called reference solution) through a trust-region search. By incorporating the predicted and reference solutions, we introduce a novel Uncertainty-based Error upper BOund (UEBO) to evaluate the uncertainty of the predicted values and fix those with high confidence. A notable feature of Apollo-MILP is the superior ability for problem reduction while preserving optimality, leading to high-quality final solutions. Experiments on commonly used benchmarks demonstrate that our proposed Apollo-MILP significantly outperforms other ML-based approaches in terms of solution quality, achieving over a 50% reduction in the solution gap.
Distinguished Quantized Guidance for Diffusion-based Sequence Recommendation
Jan 29, 2025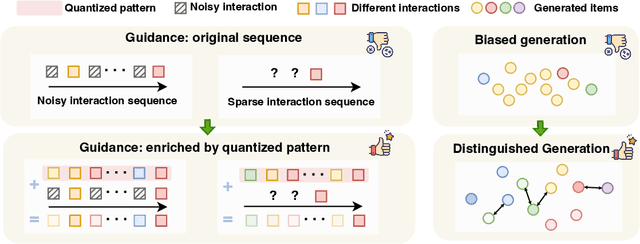
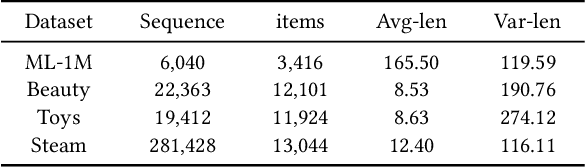
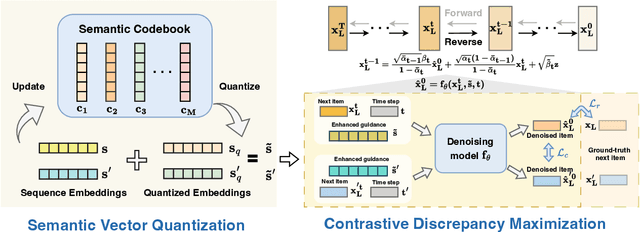
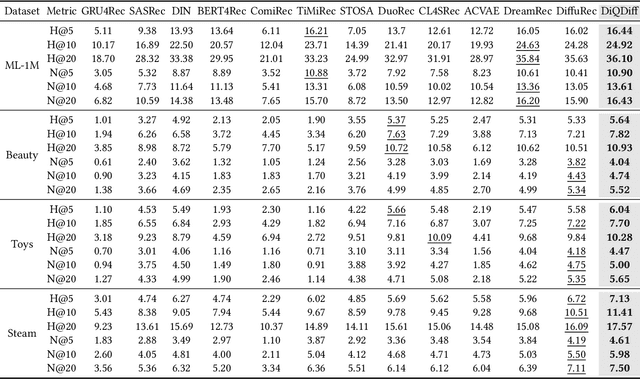
Abstract:Diffusion models (DMs) have emerged as promising approaches for sequential recommendation due to their strong ability to model data distributions and generate high-quality items. Existing work typically adds noise to the next item and progressively denoises it guided by the user's interaction sequence, generating items that closely align with user interests. However, we identify two key issues in this paradigm. First, the sequences are often heterogeneous in length and content, exhibiting noise due to stochastic user behaviors. Using such sequences as guidance may hinder DMs from accurately understanding user interests. Second, DMs are prone to data bias and tend to generate only the popular items that dominate the training dataset, thus failing to meet the personalized needs of different users. To address these issues, we propose Distinguished Quantized Guidance for Diffusion-based Sequence Recommendation (DiQDiff), which aims to extract robust guidance to understand user interests and generate distinguished items for personalized user interests within DMs. To extract robust guidance, DiQDiff introduces Semantic Vector Quantization (SVQ) to quantize sequences into semantic vectors (e.g., collaborative signals and category interests) using a codebook, which can enrich the guidance to better understand user interests. To generate distinguished items, DiQDiff personalizes the generation through Contrastive Discrepancy Maximization (CDM), which maximizes the distance between denoising trajectories using contrastive loss to prevent biased generation for different users. Extensive experiments are conducted to compare DiQDiff with multiple baseline models across four widely-used datasets. The superior recommendation performance of DiQDiff against leading approaches demonstrates its effectiveness in sequential recommendation tasks.
MILP-StuDio: MILP Instance Generation via Block Structure Decomposition
Oct 31, 2024



Abstract:Mixed-integer linear programming (MILP) is one of the most popular mathematical formulations with numerous applications. In practice, improving the performance of MILP solvers often requires a large amount of high-quality data, which can be challenging to collect. Researchers thus turn to generation techniques to generate additional MILP instances. However, existing approaches do not take into account specific block structures -- which are closely related to the problem formulations -- in the constraint coefficient matrices (CCMs) of MILPs. Consequently, they are prone to generate computationally trivial or infeasible instances due to the disruptions of block structures and thus problem formulations. To address this challenge, we propose a novel MILP generation framework, called Block Structure Decomposition (MILP-StuDio), to generate high-quality instances by preserving the block structures. Specifically, MILP-StuDio begins by identifying the blocks in CCMs and decomposing the instances into block units, which serve as the building blocks of MILP instances. We then design three operators to construct new instances by removing, substituting, and appending block units in the original instances, enabling us to generate instances with flexible sizes. An appealing feature of MILP-StuDio is its strong ability to preserve the feasibility and computational hardness of the generated instances. Experiments on the commonly-used benchmarks demonstrate that using instances generated by MILP-StuDio is able to significantly reduce over 10% of the solving time for learning-based solvers.
Invariant Graph Learning Meets Information Bottleneck for Out-of-Distribution Generalization
Aug 03, 2024Abstract:Graph out-of-distribution (OOD) generalization remains a major challenge in graph learning since graph neural networks (GNNs) often suffer from severe performance degradation under distribution shifts. Invariant learning, aiming to extract invariant features across varied distributions, has recently emerged as a promising approach for OOD generation. Despite the great success of invariant learning in OOD problems for Euclidean data (i.e., images), the exploration within graph data remains constrained by the complex nature of graphs. Existing studies, such as data augmentation or causal intervention, either suffer from disruptions to invariance during the graph manipulation process or face reliability issues due to a lack of supervised signals for causal parts. In this work, we propose a novel framework, called Invariant Graph Learning based on Information bottleneck theory (InfoIGL), to extract the invariant features of graphs and enhance models' generalization ability to unseen distributions. Specifically, InfoIGL introduces a redundancy filter to compress task-irrelevant information related to environmental factors. Cooperating with our designed multi-level contrastive learning, we maximize the mutual information among graphs of the same class in the downstream classification tasks, preserving invariant features for prediction to a great extent. An appealing feature of InfoIGL is its strong generalization ability without depending on supervised signal of invariance. Experiments on both synthetic and real-world datasets demonstrate that our method achieves state-of-the-art performance under OOD generalization for graph classification tasks. The source code is available at https://github.com/maowenyu-11/InfoIGL.
Emotion Detection through Body Gesture and Face
Jul 13, 2024Abstract:The project leverages advanced machine and deep learning techniques to address the challenge of emotion recognition by focusing on non-facial cues, specifically hands, body gestures, and gestures. Traditional emotion recognition systems mainly rely on facial expression analysis and often ignore the rich emotional information conveyed through body language. To bridge this gap, this method leverages the Aff-Wild2 and DFEW databases to train and evaluate a model capable of recognizing seven basic emotions (angry, disgust, fear, happiness, sadness, surprise, and neutral) and estimating valence and continuous scales wakeup descriptor. Leverage OpenPose for pose estimation to extract detailed body posture and posture features from images and videos. These features serve as input to state-of-the-art neural network architectures, including ResNet, and ANN for emotion classification, and fully connected layers for valence arousal regression analysis. This bifurcation strategy can solve classification and regression problems in the field of emotion recognition. The project aims to contribute to the field of affective computing by enhancing the ability of machines to interpret and respond to human emotions in a more comprehensive and nuanced way. By integrating multimodal data and cutting-edge computational models, I aspire to develop a system that not only enriches human-computer interaction but also has potential applications in areas as diverse as mental health support, educational technology, and autonomous vehicle systems.
GenoTEX: A Benchmark for Evaluating LLM-Based Exploration of Gene Expression Data in Alignment with Bioinformaticians
Jun 21, 2024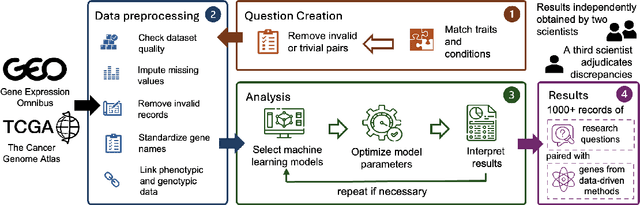
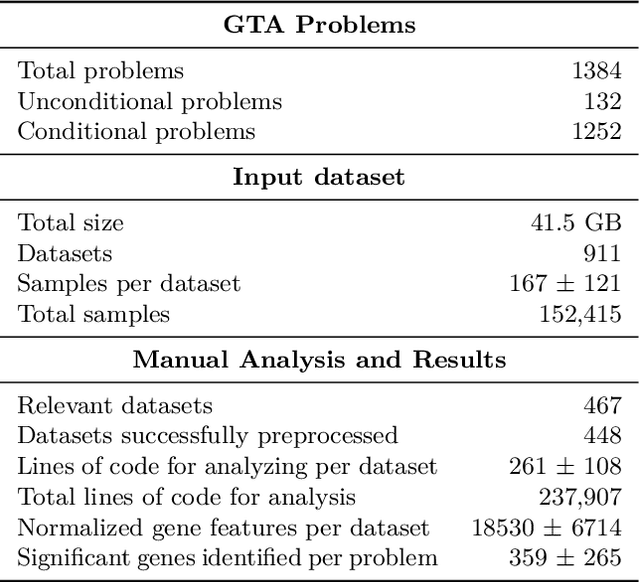
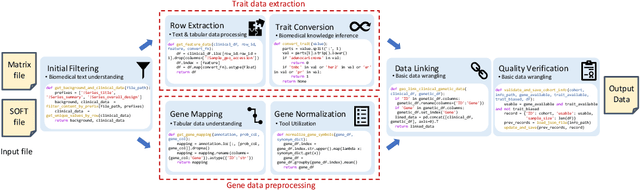
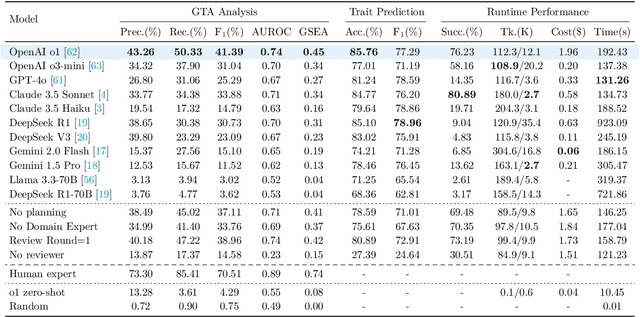
Abstract:Recent advancements in machine learning have significantly improved the identification of disease-associated genes from gene expression datasets. However, these processes often require extensive expertise and manual effort, limiting their scalability. Large Language Model (LLM)-based agents have shown promise in automating these tasks due to their increasing problem-solving abilities. To support the evaluation and development of such methods, we introduce GenoTEX, a benchmark dataset for the automatic exploration of gene expression data, involving the tasks of dataset selection, preprocessing, and statistical analysis. GenoTEX provides annotated code and results for solving a wide range of gene identification problems, in a full analysis pipeline that follows the standard of computational genomics. These annotations are curated by human bioinformaticians who carefully analyze the datasets to ensure accuracy and reliability. To provide baselines for these tasks, we present GenoAgents, a team of LLM-based agents designed with context-aware planning, iterative correction, and domain expert consultation to collaboratively explore gene datasets. Our experiments with GenoAgents demonstrate the potential of LLM-based approaches in genomics data analysis, while error analysis highlights the challenges and areas for future improvement. We propose GenoTEX as a promising resource for benchmarking and enhancing AI-driven methods for genomics data analysis. We make our benchmark publicly available at \url{https://github.com/Liu-Hy/GenoTex}.
Deep Symbolic Optimization for Combinatorial Optimization: Accelerating Node Selection by Discovering Potential Heuristics
Jun 14, 2024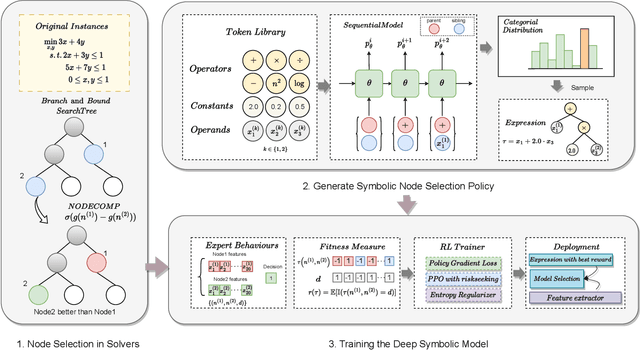
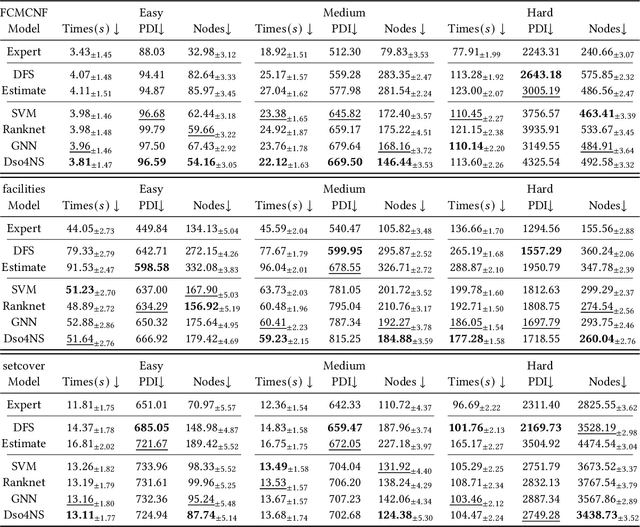
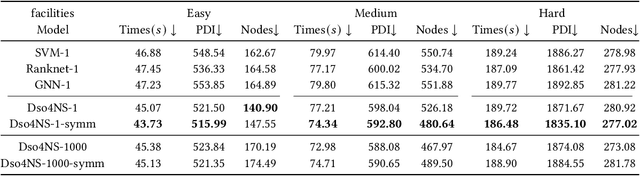
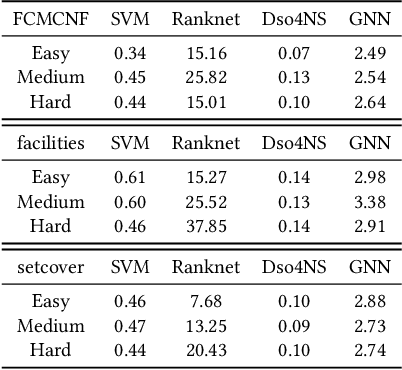
Abstract:Combinatorial optimization (CO) is one of the most fundamental mathematical models in real-world applications. Traditional CO solvers, such as Branch-and-Bound (B&B) solvers, heavily rely on expert-designed heuristics, which are reliable but require substantial manual tuning. Recent studies have leveraged deep learning (DL) models as an alternative to capture rich feature patterns for improved performance on GPU machines. Nonetheless, the drawbacks of high training and inference costs, as well as limited interpretability, severely hinder the adoption of DL methods in real-world applications. To address these challenges, we propose a novel deep symbolic optimization learning framework that combines their advantages. Specifically, we focus on the node selection module within B&B solvers -- namely, deep symbolic optimization for node selection (Dso4NS). With data-driven approaches, Dso4NS guides the search for mathematical expressions within the high-dimensional discrete symbolic space and then incorporates the highest-performing mathematical expressions into a solver. The data-driven model captures the rich feature information in the input data and generates symbolic expressions, while the expressions deployed in solvers enable fast inference with high interpretability. Experiments demonstrate the effectiveness of Dso4NS in learning high-quality expressions, outperforming existing approaches on a CPU machine. Encouragingly, the learned CPU-based policies consistently achieve performance comparable to state-of-the-art GPU-based approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge