Yuxuan Cheng
TED-LaST: Towards Robust Backdoor Defense Against Adaptive Attacks
Jun 12, 2025Abstract:Deep Neural Networks (DNNs) are vulnerable to backdoor attacks, where attackers implant hidden triggers during training to maliciously control model behavior. Topological Evolution Dynamics (TED) has recently emerged as a powerful tool for detecting backdoor attacks in DNNs. However, TED can be vulnerable to backdoor attacks that adaptively distort topological representation distributions across network layers. To address this limitation, we propose TED-LaST (Topological Evolution Dynamics against Laundry, Slow release, and Target mapping attack strategies), a novel defense strategy that enhances TED's robustness against adaptive attacks. TED-LaST introduces two key innovations: label-supervised dynamics tracking and adaptive layer emphasis. These enhancements enable the identification of stealthy threats that evade traditional TED-based defenses, even in cases of inseparability in topological space and subtle topological perturbations. We review and classify data poisoning tricks in state-of-the-art adaptive attacks and propose enhanced adaptive attack with target mapping, which can dynamically shift malicious tasks and fully leverage the stealthiness that adaptive attacks possess. Our comprehensive experiments on multiple datasets (CIFAR-10, GTSRB, and ImageNet100) and model architectures (ResNet20, ResNet101) show that TED-LaST effectively counteracts sophisticated backdoors like Adap-Blend, Adapt-Patch, and the proposed enhanced adaptive attack. TED-LaST sets a new benchmark for robust backdoor detection, substantially enhancing DNN security against evolving threats.
A4-Unet: Deformable Multi-Scale Attention Network for Brain Tumor Segmentation
Dec 08, 2024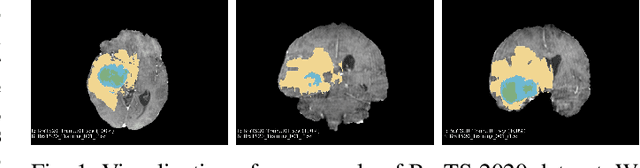
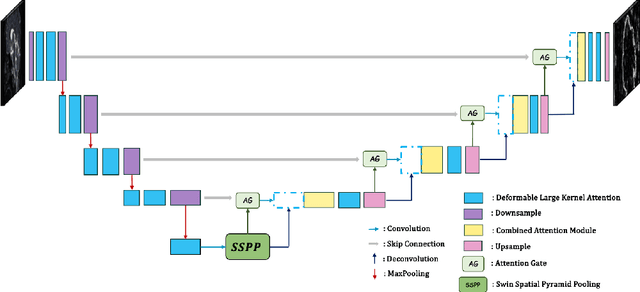
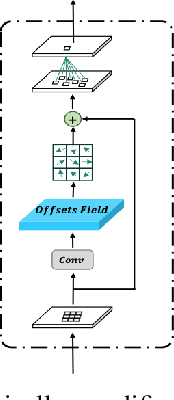
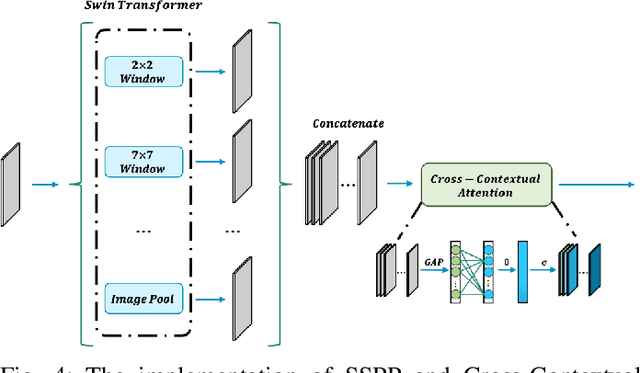
Abstract:Brain tumor segmentation models have aided diagnosis in recent years. However, they face MRI complexity and variability challenges, including irregular shapes and unclear boundaries, leading to noise, misclassification, and incomplete segmentation, thereby limiting accuracy. To address these issues, we adhere to an outstanding Convolutional Neural Networks (CNNs) design paradigm and propose a novel network named A4-Unet. In A4-Unet, Deformable Large Kernel Attention (DLKA) is incorporated in the encoder, allowing for improved capture of multi-scale tumors. Swin Spatial Pyramid Pooling (SSPP) with cross-channel attention is employed in a bottleneck further to study long-distance dependencies within images and channel relationships. To enhance accuracy, a Combined Attention Module (CAM) with Discrete Cosine Transform (DCT) orthogonality for channel weighting and convolutional element-wise multiplication is introduced for spatial weighting in the decoder. Attention gates (AG) are added in the skip connection to highlight the foreground while suppressing irrelevant background information. The proposed network is evaluated on three authoritative MRI brain tumor benchmarks and a proprietary dataset, and it achieves a 94.4% Dice score on the BraTS 2020 dataset, thereby establishing multiple new state-of-the-art benchmarks. The code is available here: https://github.com/WendyWAAAAANG/A4-Unet.
Attention-Guided Perturbation for Consistency Regularization in Semi-Supervised Medical Image Segmentation
Oct 16, 2024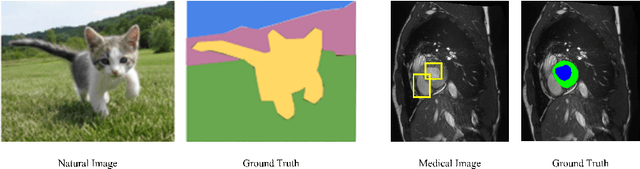
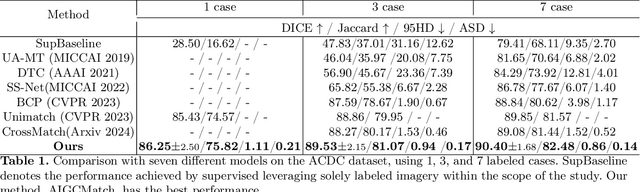
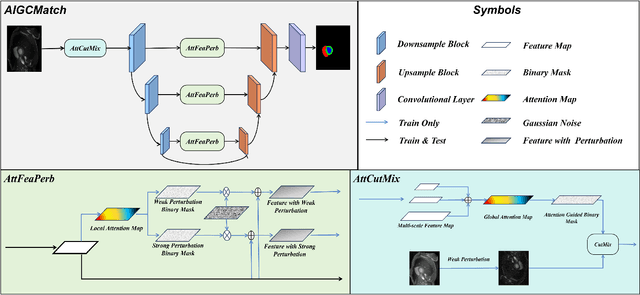
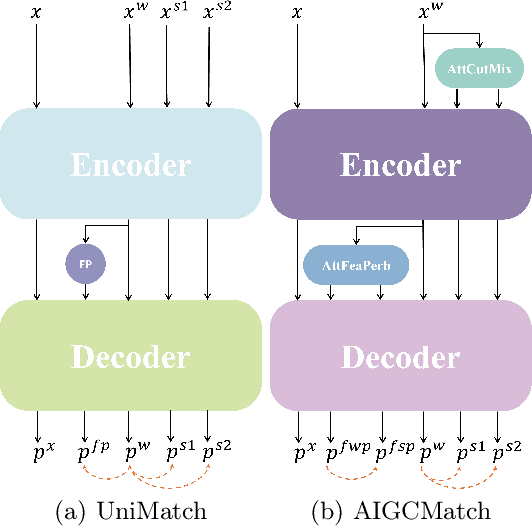
Abstract:Medical image segmentation is a pivotal step in diagnostic and therapeutic processes. However, the acquisition of high-quality annotated data is often constrained by scarcity and cost. Semi-supervised learning offers a promising approach to enhance model performance by using unlabeled data. While consistency regularization is a prevalent method in semi-supervised image segmentation, there is a dearth of research on perturbation strategies tailored for semi-supervised medical image segmentation tasks. This paper introduces an attention-guided perturbation strategy for semi-supervised consistency regularization in the context of medical image segmentation. We add the perturbation based on the attention from the model in the image and feature level to achieve consistency regularization. The method is adept at accommodating the intricate structures and high-dimensional semantics inherent in medical images, thereby enhancing the performance of semi-supervised segmentation tasks. Our method achieved state-of-the-art results on benchmark datasets, including a 90.4\% Dice score on the ACDC dataset in the 7-case scenario.
Attention-Guided Perturbation for Unsupervised Image Anomaly Detection
Aug 14, 2024Abstract:Reconstruction-based methods have significantly advanced modern unsupervised anomaly detection. However, the strong capacity of neural networks often violates the underlying assumptions by reconstructing abnormal samples well. To alleviate this issue, we present a simple yet effective reconstruction framework named Attention-Guided Pertuation Network (AGPNet), which learns to add perturbation noise with an attention mask, for accurate unsupervised anomaly detection. Specifically, it consists of two branches, \ie, a plain reconstruction branch and an auxiliary attention-based perturbation branch. The reconstruction branch is simply a plain reconstruction network that learns to reconstruct normal samples, while the auxiliary branch aims to produce attention masks to guide the noise perturbation process for normal samples from easy to hard. By doing so, we are expecting to synthesize hard yet more informative anomalies for training, which enable the reconstruction branch to learn important inherent normal patterns both comprehensively and efficiently. Extensive experiments are conducted on three popular benchmarks covering MVTec-AD, VisA, and MVTec-3D, and show that our framework obtains leading anomaly detection performance under various setups including few-shot, one-class, and multi-class setups.
SimpleFusion: A Simple Fusion Framework for Infrared and Visible Images
Jun 27, 2024Abstract:Integrating visible and infrared images into one high-quality image, also known as visible and infrared image fusion, is a challenging yet critical task for many downstream vision tasks. Most existing works utilize pretrained deep neural networks or design sophisticated frameworks with strong priors for this task, which may be unsuitable or lack flexibility. This paper presents SimpleFusion, a simple yet effective framework for visible and infrared image fusion. Our framework follows the decompose-and-fusion paradigm, where the visible and the infrared images are decomposed into reflectance and illumination components via Retinex theory and followed by the fusion of these corresponding elements. The whole framework is designed with two plain convolutional neural networks without downsampling, which can perform image decomposition and fusion efficiently. Moreover, we introduce decomposition loss and a detail-to-semantic loss to preserve the complementary information between the two modalities for fusion. We conduct extensive experiments on the challenging benchmarks, verifying the superiority of our method over previous state-of-the-arts. Code is available at \href{https://github.com/hxwxss/SimpleFusion-A-Simple-Fusion-Framework-for-Infrared-and-Visible-Images}{https://github.com/hxwxss/SimpleFusion-A-Simple-Fusion-Framework-for-Infrared-and-Visible-Images}
Toward a Team of AI-made Scientists for Scientific Discovery from Gene Expression Data
Feb 21, 2024



Abstract:Machine learning has emerged as a powerful tool for scientific discovery, enabling researchers to extract meaningful insights from complex datasets. For instance, it has facilitated the identification of disease-predictive genes from gene expression data, significantly advancing healthcare. However, the traditional process for analyzing such datasets demands substantial human effort and expertise for the data selection, processing, and analysis. To address this challenge, we introduce a novel framework, a Team of AI-made Scientists (TAIS), designed to streamline the scientific discovery pipeline. TAIS comprises simulated roles, including a project manager, data engineer, and domain expert, each represented by a Large Language Model (LLM). These roles collaborate to replicate the tasks typically performed by data scientists, with a specific focus on identifying disease-predictive genes. Furthermore, we have curated a benchmark dataset to assess TAIS's effectiveness in gene identification, demonstrating our system's potential to significantly enhance the efficiency and scope of scientific exploration. Our findings represent a solid step towards automating scientific discovery through large language models.
Beyond Pixels: Exploring Human-Readable SVG Generation for Simple Images with Vision Language Models
Nov 27, 2023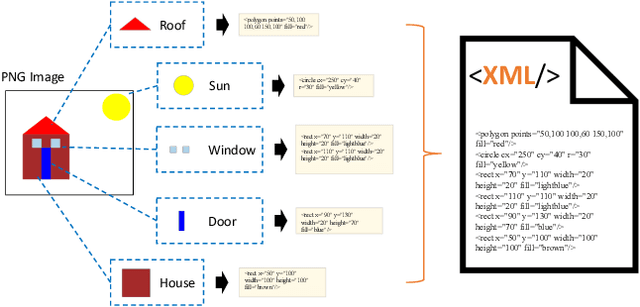
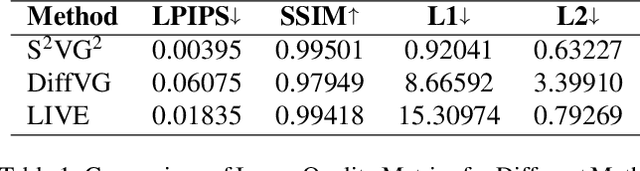
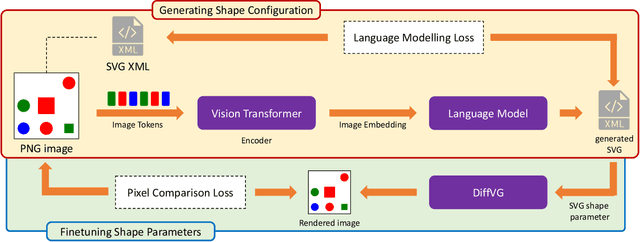
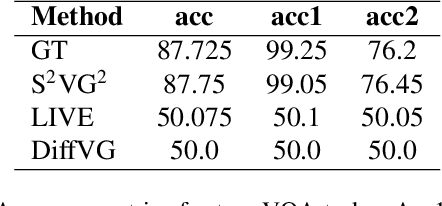
Abstract:In the field of computer graphics, the use of vector graphics, particularly Scalable Vector Graphics (SVG), represents a notable development from traditional pixel-based imagery. SVGs, with their XML-based format, are distinct in their ability to directly and explicitly represent visual elements such as shape, color, and path. This direct representation facilitates a more accurate and logical depiction of graphical elements, enhancing reasoning and interpretability. Recognizing the potential of SVGs, the machine learning community has introduced multiple methods for image vectorization. However, transforming images into SVG format while retaining the relational properties and context of the original scene remains a key challenge. Most vectorization methods often yield SVGs that are overly complex and not easily interpretable. In response to this challenge, we introduce our method, Simple-SVG-Generation (S\textsuperscript{2}VG\textsuperscript{2}). Our method focuses on producing SVGs that are both accurate and simple, aligning with human readability and understanding. With simple images, we evaluate our method with reasoning tasks together with advanced language models, the results show a clear improvement over previous SVG generation methods. We also conducted surveys for human evaluation on the readability of our generated SVGs, the results also favor our methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge