A4-Unet: Deformable Multi-Scale Attention Network for Brain Tumor Segmentation
Paper and Code
Dec 08, 2024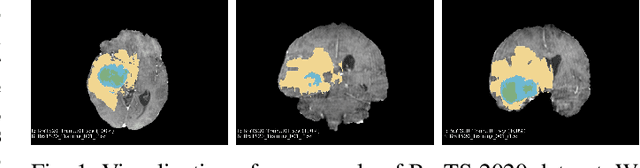
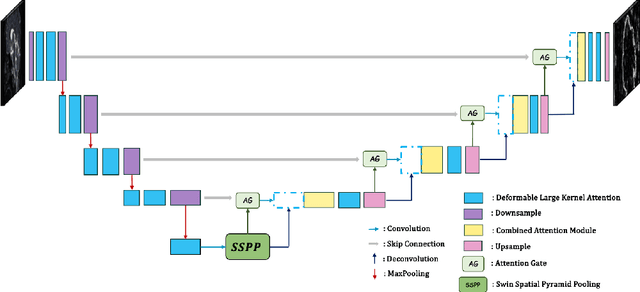
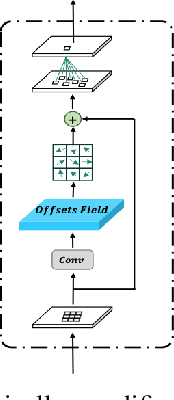
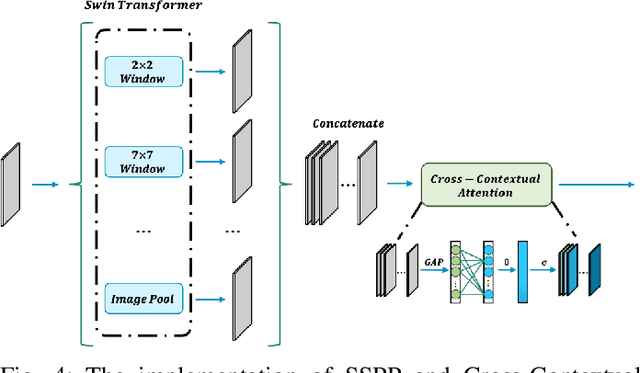
Brain tumor segmentation models have aided diagnosis in recent years. However, they face MRI complexity and variability challenges, including irregular shapes and unclear boundaries, leading to noise, misclassification, and incomplete segmentation, thereby limiting accuracy. To address these issues, we adhere to an outstanding Convolutional Neural Networks (CNNs) design paradigm and propose a novel network named A4-Unet. In A4-Unet, Deformable Large Kernel Attention (DLKA) is incorporated in the encoder, allowing for improved capture of multi-scale tumors. Swin Spatial Pyramid Pooling (SSPP) with cross-channel attention is employed in a bottleneck further to study long-distance dependencies within images and channel relationships. To enhance accuracy, a Combined Attention Module (CAM) with Discrete Cosine Transform (DCT) orthogonality for channel weighting and convolutional element-wise multiplication is introduced for spatial weighting in the decoder. Attention gates (AG) are added in the skip connection to highlight the foreground while suppressing irrelevant background information. The proposed network is evaluated on three authoritative MRI brain tumor benchmarks and a proprietary dataset, and it achieves a 94.4% Dice score on the BraTS 2020 dataset, thereby establishing multiple new state-of-the-art benchmarks. The code is available here: https://github.com/WendyWAAAAANG/A4-Unet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge