Lisheng Wang
SegRap2025: A Benchmark of Gross Tumor Volume and Lymph Node Clinical Target Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Jan 28, 2026Abstract:Accurate delineation of Gross Tumor Volume (GTV), Lymph Node Clinical Target Volume (LN CTV), and Organ-at-Risk (OAR) from Computed Tomography (CT) scans is essential for precise radiotherapy planning in Nasopharyngeal Carcinoma (NPC). Building upon SegRap2023, which focused on OAR and GTV segmentation using single-center paired non-contrast CT (ncCT) and contrast-enhanced CT (ceCT) scans, the SegRap2025 challenge aims to enhance the generalizability and robustness of segmentation models across imaging centers and modalities. SegRap2025 comprises two tasks: Task01 addresses GTV segmentation using paired CT from the SegRap2023 dataset, with an additional external testing set to evaluate cross-center generalization, and Task02 focuses on LN CTV segmentation using multi-center training data and an unseen external testing set, where each case contains paired CT scans or a single modality, emphasizing both cross-center and cross-modality robustness. This paper presents the challenge setup and provides a comprehensive analysis of the solutions submitted by ten participating teams. For GTV segmentation task, the top-performing models achieved average Dice Similarity Coefficient (DSC) of 74.61% and 56.79% on the internal and external testing cohorts, respectively. For LN CTV segmentation task, the highest average DSC values reached 60.24%, 60.50%, and 57.23% on paired CT, ceCT-only, and ncCT-only subsets, respectively. SegRap2025 establishes a large-scale multi-center, multi-modality benchmark for evaluating the generalization and robustness in radiotherapy target segmentation, providing valuable insights toward clinically applicable automated radiotherapy planning systems. The benchmark is available at: https://hilab-git.github.io/SegRap2025_Challenge.
Unsupervised Anomaly Detection in Brain MRI via Disentangled Anatomy Learning
Dec 26, 2025Abstract:Detection of various lesions in brain MRI is clinically critical, but challenging due to the diversity of lesions and variability in imaging conditions. Current unsupervised learning methods detect anomalies mainly through reconstructing abnormal images into pseudo-healthy images (PHIs) by normal samples learning and then analyzing differences between images. However, these unsupervised models face two significant limitations: restricted generalizability to multi-modality and multi-center MRIs due to their reliance on the specific imaging information in normal training data, and constrained performance due to abnormal residuals propagated from input images to reconstructed PHIs. To address these limitations, two novel modules are proposed, forming a new PHI reconstruction framework. Firstly, the disentangled representation module is proposed to improve generalizability by decoupling brain MRI into imaging information and essential imaging-invariant anatomical images, ensuring that the reconstruction focuses on the anatomy. Specifically, brain anatomical priors and a differentiable one-hot encoding operator are introduced to constrain the disentanglement results and enhance the disentanglement stability. Secondly, the edge-to-image restoration module is designed to reconstruct high-quality PHIs by restoring the anatomical representation from the high-frequency edge information of anatomical images, and then recoupling the disentangled imaging information. This module not only suppresses abnormal residuals in PHI by reducing abnormal pixels input through edge-only input, but also effectively reconstructs normal regions using the preserved structural details in the edges. Evaluated on nine public datasets (4,443 patients' MRIs from multiple centers), our method outperforms 17 SOTA methods, achieving absolute improvements of +18.32% in AP and +13.64% in DSC.
* Accepted by Medical Image Analysis (2025)
crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023
Jun 13, 2025Abstract:The cross-Modality Domain Adaptation (crossMoDA) challenge series, initiated in 2021 in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), focuses on unsupervised cross-modality segmentation, learning from contrast-enhanced T1 (ceT1) and transferring to T2 MRI. The task is an extreme example of domain shift chosen to serve as a meaningful and illustrative benchmark. From a clinical application perspective, it aims to automate Vestibular Schwannoma (VS) and cochlea segmentation on T2 scans for more cost-effective VS management. Over time, the challenge objectives have evolved to enhance its clinical relevance. The challenge evolved from using single-institutional data and basic segmentation in 2021 to incorporating multi-institutional data and Koos grading in 2022, and by 2023, it included heterogeneous routine data and sub-segmentation of intra- and extra-meatal tumour components. In this work, we report the findings of the 2022 and 2023 editions and perform a retrospective analysis of the challenge progression over the years. The observations from the successive challenge contributions indicate that the number of outliers decreases with an expanding dataset. This is notable since the diversity of scanning protocols of the datasets concurrently increased. The winning approach of the 2023 edition reduced the number of outliers on the 2021 and 2022 testing data, demonstrating how increased data heterogeneity can enhance segmentation performance even on homogeneous data. However, the cochlea Dice score declined in 2023, likely due to the added complexity from tumour sub-annotations affecting overall segmentation performance. While progress is still needed for clinically acceptable VS segmentation, the plateauing performance suggests that a more challenging cross-modal task may better serve future benchmarking.
Image Translation-Based Unsupervised Cross-Modality Domain Adaptation for Medical Image Segmentation
Feb 21, 2025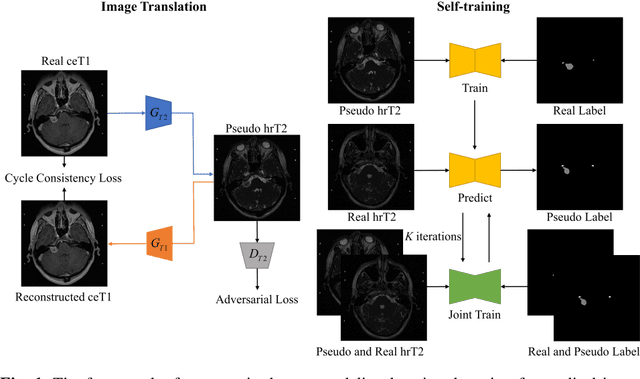

Abstract:Supervised deep learning usually faces more challenges in medical images than in natural images. Since annotations in medical images require the expertise of doctors and are more time-consuming and expensive. Thus, some researchers turn to unsupervised learning methods, which usually face inevitable performance drops. In addition, medical images may have been acquired at different medical centers with different scanners and under different image acquisition protocols, so the modalities of the medical images are often inconsistent. This modality difference (domain shift) also reduces the applicability of deep learning methods. In this regard, we propose an unsupervised crossmodality domain adaptation method based on image translation by transforming the source modality image with annotation into the unannotated target modality and using its annotation to achieve supervised learning of the target modality. In addition, the subtle differences between translated pseudo images and real images are overcome by self-training methods to further improve the task performance of deep learning. The proposed method showed mean Dice Similarity Coefficient (DSC) and Average Symmetric Surface Distance (ASSD) of $0.8351 \pm 0.1152$ and $1.6712 \pm 2.1948$ for vestibular schwannoma (VS), $0.8098 \pm 0.0233$ and $0.2317 \pm 0.1577$ for cochlea on the VS and cochlea segmentation task of the Cross-Modality Domain Adaptation (crossMoDA 2022) challenge validation phase leaderboard.
Multi-Class Segmentation of Aortic Branches and Zones in Computed Tomography Angiography: The AortaSeg24 Challenge
Feb 07, 2025



Abstract:Multi-class segmentation of the aorta in computed tomography angiography (CTA) scans is essential for diagnosing and planning complex endovascular treatments for patients with aortic dissections. However, existing methods reduce aortic segmentation to a binary problem, limiting their ability to measure diameters across different branches and zones. Furthermore, no open-source dataset is currently available to support the development of multi-class aortic segmentation methods. To address this gap, we organized the AortaSeg24 MICCAI Challenge, introducing the first dataset of 100 CTA volumes annotated for 23 clinically relevant aortic branches and zones. This dataset was designed to facilitate both model development and validation. The challenge attracted 121 teams worldwide, with participants leveraging state-of-the-art frameworks such as nnU-Net and exploring novel techniques, including cascaded models, data augmentation strategies, and custom loss functions. We evaluated the submitted algorithms using the Dice Similarity Coefficient (DSC) and Normalized Surface Distance (NSD), highlighting the approaches adopted by the top five performing teams. This paper presents the challenge design, dataset details, evaluation metrics, and an in-depth analysis of the top-performing algorithms. The annotated dataset, evaluation code, and implementations of the leading methods are publicly available to support further research. All resources can be accessed at https://aortaseg24.grand-challenge.org.
DPCL-Diff: The Temporal Knowledge Graph Reasoning based on Graph Node Diffusion Model with Dual-Domain Periodic Contrastive Learning
Nov 03, 2024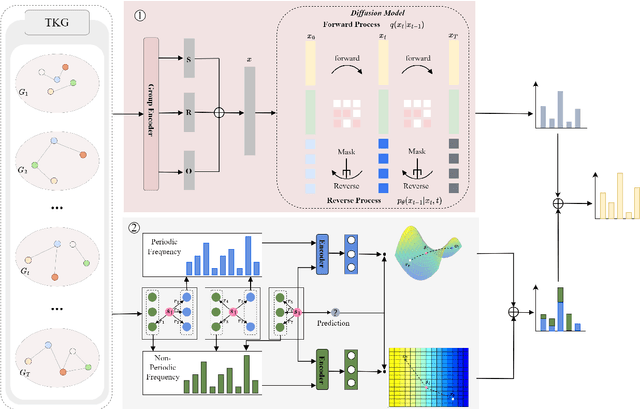
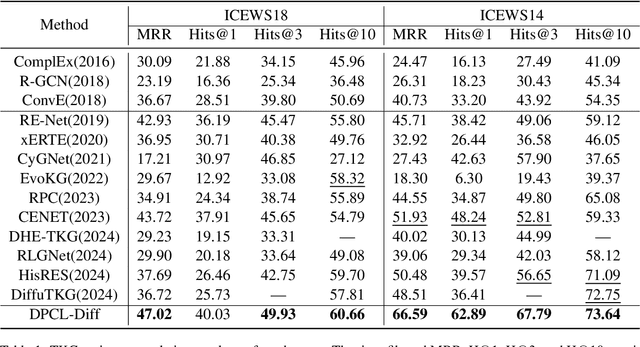
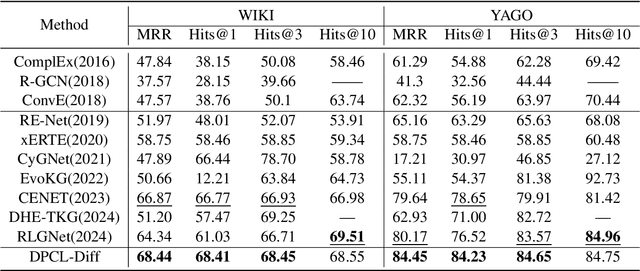
Abstract:Temporal knowledge graph (TKG) reasoning that infers future missing facts is an essential and challenging task. Predicting future events typically relies on closely related historical facts, yielding more accurate results for repetitive or periodic events. However, for future events with sparse historical interactions, the effectiveness of this method, which focuses on leveraging high-frequency historical information, diminishes. Recently, the capabilities of diffusion models in image generation have opened new opportunities for TKG reasoning. Therefore, we propose a graph node diffusion model with dual-domain periodic contrastive learning (DPCL-Diff). Graph node diffusion model (GNDiff) introduces noise into sparsely related events to simulate new events, generating high-quality data that better conforms to the actual distribution. This generative mechanism significantly enhances the model's ability to reason about new events. Additionally, the dual-domain periodic contrastive learning (DPCL) maps periodic and non-periodic event entities to Poincar\'e and Euclidean spaces, leveraging their characteristics to distinguish similar periodic events effectively. Experimental results on four public datasets demonstrate that DPCL-Diff significantly outperforms state-of-the-art TKG models in event prediction, demonstrating our approach's effectiveness. This study also investigates the combined effectiveness of GNDiff and DPCL in TKG tasks.
SegRap2023: A Benchmark of Organs-at-Risk and Gross Tumor Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Dec 15, 2023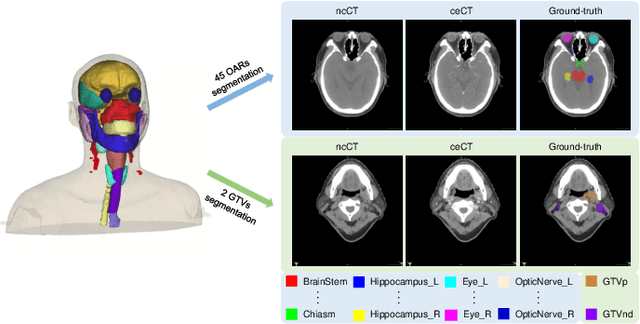
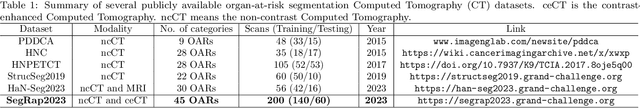
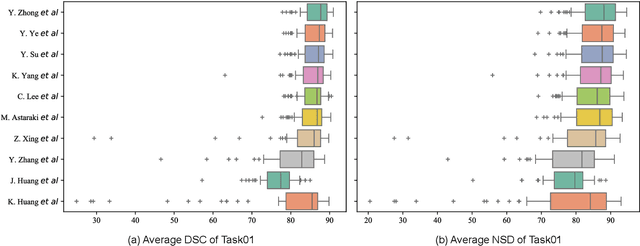
Abstract:Radiation therapy is a primary and effective NasoPharyngeal Carcinoma (NPC) treatment strategy. The precise delineation of Gross Tumor Volumes (GTVs) and Organs-At-Risk (OARs) is crucial in radiation treatment, directly impacting patient prognosis. Previously, the delineation of GTVs and OARs was performed by experienced radiation oncologists. Recently, deep learning has achieved promising results in many medical image segmentation tasks. However, for NPC OARs and GTVs segmentation, few public datasets are available for model development and evaluation. To alleviate this problem, the SegRap2023 challenge was organized in conjunction with MICCAI2023 and presented a large-scale benchmark for OAR and GTV segmentation with 400 Computed Tomography (CT) scans from 200 NPC patients, each with a pair of pre-aligned non-contrast and contrast-enhanced CT scans. The challenge's goal was to segment 45 OARs and 2 GTVs from the paired CT scans. In this paper, we detail the challenge and analyze the solutions of all participants. The average Dice similarity coefficient scores for all submissions ranged from 76.68\% to 86.70\%, and 70.42\% to 73.44\% for OARs and GTVs, respectively. We conclude that the segmentation of large-size OARs is well-addressed, and more efforts are needed for GTVs and small-size or thin-structure OARs. The benchmark will remain publicly available here: https://segrap2023.grand-challenge.org
Intergrated Segmentation and Detection Models for Dentex Challenge 2023
Sep 04, 2023



Abstract:Dental panoramic x-rays are commonly used in dental diagnosing. With the development of deep learning, auto detection of diseases from dental panoramic x-rays can help dentists to diagnose diseases more efficiently.The Dentex Challenge 2023 is a competition for automatic detection of abnormal teeth along with their enumeration ids from dental panoramic x-rays. In this paper, we propose a method integrating segmentation and detection models to detect abnormal teeth as well as obtain their enumeration ids.Our codes are available at https://github.com/xyzlancehe/DentexSegAndDet.
Inferior Alveolar Nerve Segmentation in CBCT images using Connectivity-Based Selective Re-training
Aug 18, 2023


Abstract:Inferior Alveolar Nerve (IAN) canal detection in CBCT is an important step in many dental and maxillofacial surgery applications to prevent irreversible damage to the nerve during the procedure.The ToothFairy2023 Challenge aims to establish a 3D maxillofacial dataset consisting of all sparse labels and partial dense labels, and improve the ability of automatic IAN segmentation. In this work, in order to avoid the negative impact brought by sparse labeling, we transform the mixed supervised problem into a semi-supervised problem. Inspired by self-training via pseudo labeling, we propose a selective re-training framework based on IAN connectivity. Our method is quantitatively evaluated on the ToothFairy verification cases, achieving the dice similarity coefficient (DSC) of 0.7956, and 95\% hausdorff distance (HD95) of 4.4905, and wining the champion in the competition. Code is available at https://github.com/GaryNico517/SSL-IAN-Retraining.
The KiTS21 Challenge: Automatic segmentation of kidneys, renal tumors, and renal cysts in corticomedullary-phase CT
Jul 05, 2023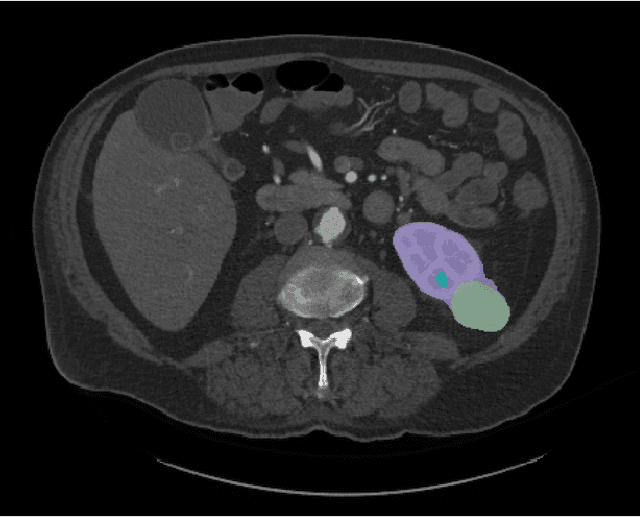
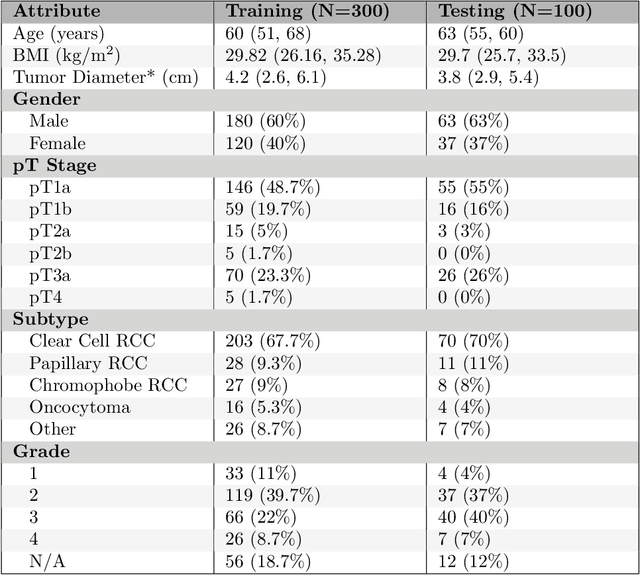
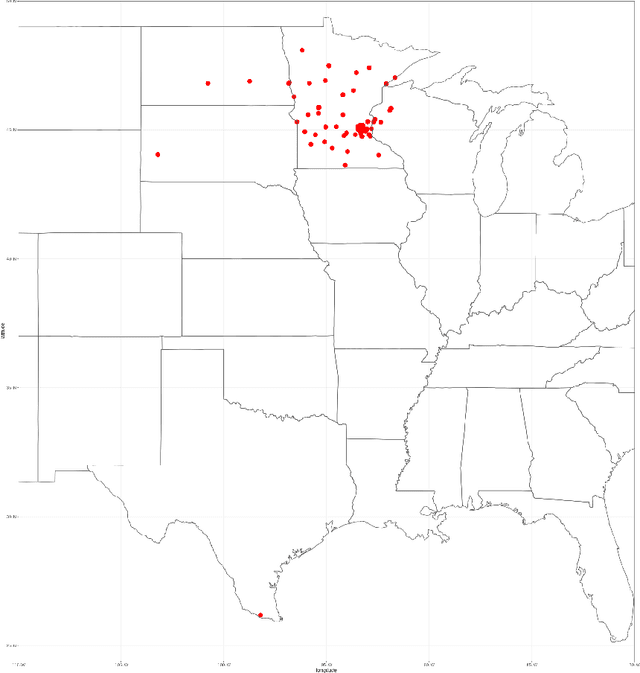
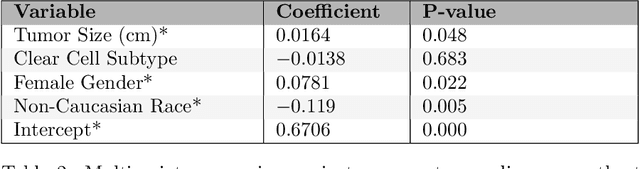
Abstract:This paper presents the challenge report for the 2021 Kidney and Kidney Tumor Segmentation Challenge (KiTS21) held in conjunction with the 2021 international conference on Medical Image Computing and Computer Assisted Interventions (MICCAI). KiTS21 is a sequel to its first edition in 2019, and it features a variety of innovations in how the challenge was designed, in addition to a larger dataset. A novel annotation method was used to collect three separate annotations for each region of interest, and these annotations were performed in a fully transparent setting using a web-based annotation tool. Further, the KiTS21 test set was collected from an outside institution, challenging participants to develop methods that generalize well to new populations. Nonetheless, the top-performing teams achieved a significant improvement over the state of the art set in 2019, and this performance is shown to inch ever closer to human-level performance. An in-depth meta-analysis is presented describing which methods were used and how they faired on the leaderboard, as well as the characteristics of which cases generally saw good performance, and which did not. Overall KiTS21 facilitated a significant advancement in the state of the art in kidney tumor segmentation, and provides useful insights that are applicable to the field of semantic segmentation as a whole.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge