Teng Zhang
WorldRFT: Latent World Model Planning with Reinforcement Fine-Tuning for Autonomous Driving
Dec 22, 2025Abstract:Latent World Models enhance scene representation through temporal self-supervised learning, presenting a perception annotation-free paradigm for end-to-end autonomous driving. However, the reconstruction-oriented representation learning tangles perception with planning tasks, leading to suboptimal optimization for planning. To address this challenge, we propose WorldRFT, a planning-oriented latent world model framework that aligns scene representation learning with planning via a hierarchical planning decomposition and local-aware interactive refinement mechanism, augmented by reinforcement learning fine-tuning (RFT) to enhance safety-critical policy performance. Specifically, WorldRFT integrates a vision-geometry foundation model to improve 3D spatial awareness, employs hierarchical planning task decomposition to guide representation optimization, and utilizes local-aware iterative refinement to derive a planning-oriented driving policy. Furthermore, we introduce Group Relative Policy Optimization (GRPO), which applies trajectory Gaussianization and collision-aware rewards to fine-tune the driving policy, yielding systematic improvements in safety. WorldRFT achieves state-of-the-art (SOTA) performance on both open-loop nuScenes and closed-loop NavSim benchmarks. On nuScenes, it reduces collision rates by 83% (0.30% -> 0.05%). On NavSim, using camera-only sensors input, it attains competitive performance with the LiDAR-based SOTA method DiffusionDrive (87.8 vs. 88.1 PDMS).
Benchmarking neural surrogates on realistic spatiotemporal multiphysics flows
Dec 21, 2025Abstract:Predicting multiphysics dynamics is computationally expensive and challenging due to the severe coupling of multi-scale, heterogeneous physical processes. While neural surrogates promise a paradigm shift, the field currently suffers from an "illusion of mastery", as repeatedly emphasized in top-tier commentaries: existing evaluations overly rely on simplified, low-dimensional proxies, which fail to expose the models' inherent fragility in realistic regimes. To bridge this critical gap, we present REALM (REalistic AI Learning for Multiphysics), a rigorous benchmarking framework designed to test neural surrogates on challenging, application-driven reactive flows. REALM features 11 high-fidelity datasets spanning from canonical multiphysics problems to complex propulsion and fire safety scenarios, alongside a standardized end-to-end training and evaluation protocol that incorporates multiphysics-aware preprocessing and a robust rollout strategy. Using this framework, we systematically benchmark over a dozen representative surrogate model families, including spectral operators, convolutional models, Transformers, pointwise operators, and graph/mesh networks, and identify three robust trends: (i) a scaling barrier governed jointly by dimensionality, stiffness, and mesh irregularity, leading to rapidly growing rollout errors; (ii) performance primarily controlled by architectural inductive biases rather than parameter count; and (iii) a persistent gap between nominal accuracy metrics and physically trustworthy behavior, where models with high correlations still miss key transient structures and integral quantities. Taken together, REALM exposes the limits of current neural surrogates on realistic multiphysics flows and offers a rigorous testbed to drive the development of next-generation physics-aware architectures.
A Dual-Feature Extractor Framework for Accurate Back Depth and Spine Morphology Estimation from Monocular RGB Images
Jul 30, 2025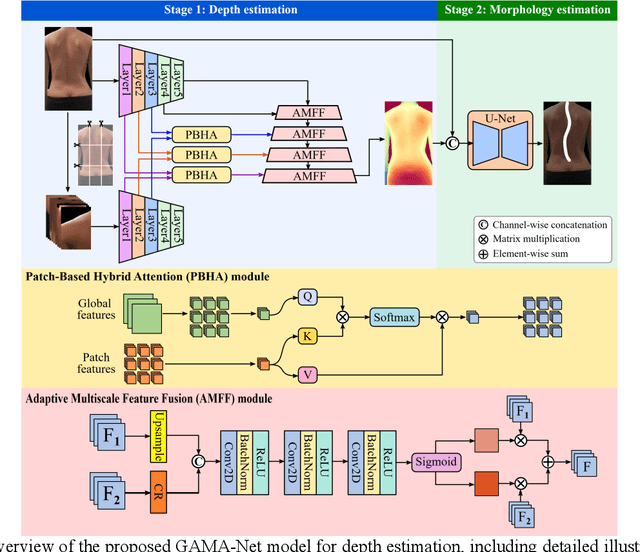

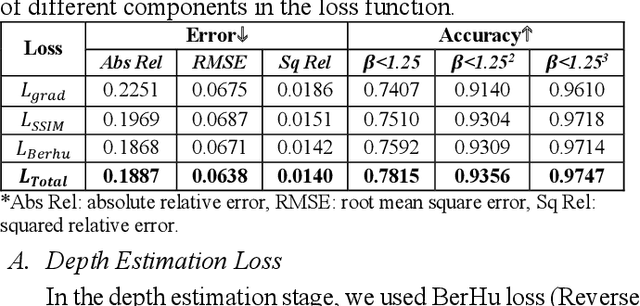
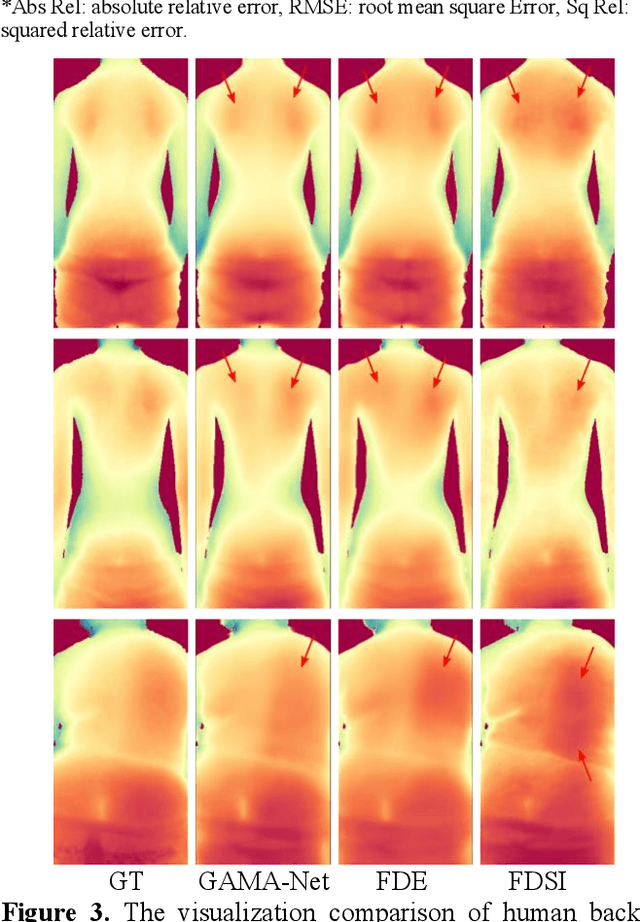
Abstract:Scoliosis is a prevalent condition that impacts both physical health and appearance, with adolescent idiopathic scoliosis (AIS) being the most common form. Currently, the main AIS assessment tool, X-rays, poses significant limitations, including radiation exposure and limited accessibility in poor and remote areas. To address this problem, the current solutions are using RGB images to analyze spine morphology. However, RGB images are highly susceptible to environmental factors, such as lighting conditions, compromising model stability and generalizability. Therefore, in this study, we propose a novel pipeline to accurately estimate the depth information of the unclothed back, compensating for the limitations of 2D information, and then estimate spine morphology by integrating both depth and surface information. To capture the subtle depth variations of the back surface with precision, we design an adaptive multiscale feature learning network named Grid-Aware Multiscale Adaptive Network (GAMA-Net). This model uses dual encoders to extract both patch-level and global features, which are then interacted by the Patch-Based Hybrid Attention (PBHA) module. The Adaptive Multiscale Feature Fusion (AMFF) module is used to dynamically fuse information in the decoder. As a result, our depth estimation model achieves remarkable accuracy across three different evaluation metrics, with scores of nearly 78.2%, 93.6%, and 97.5%, respectively. To further validate the effectiveness of the predicted depth, we integrate both surface and depth information for spine morphology estimation. This integrated approach enhances the accuracy of spine curve generation, achieving an impressive performance of up to 97%.
NGA: Non-autoregressive Generative Auction with Global Externalities for Advertising Systems
Jun 06, 2025


Abstract:Online advertising auctions are fundamental to internet commerce, demanding solutions that not only maximize revenue but also ensure incentive compatibility, high-quality user experience, and real-time efficiency. While recent learning-based auction frameworks have improved context modeling by capturing intra-list dependencies among ads, they remain limited in addressing global externalities and often suffer from inefficiencies caused by sequential processing. In this work, we introduce the Non-autoregressive Generative Auction with global externalities (NGA), a novel end-to-end framework designed for industrial online advertising. NGA explicitly models global externalities by jointly capturing the relationships among ads as well as the effects of adjacent organic content. To further enhance efficiency, NGA utilizes a non-autoregressive, constraint-based decoding strategy and a parallel multi-tower evaluator for unified list-wise reward and payment computation. Extensive offline experiments and large-scale online A/B testing on commercial advertising platforms demonstrate that NGA consistently outperforms existing methods in both effectiveness and efficiency.
Beyond Cascaded Architectures: An End-to-end Generative Framework for Industrial Advertising
May 26, 2025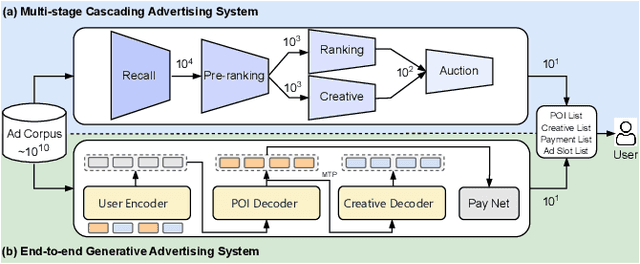
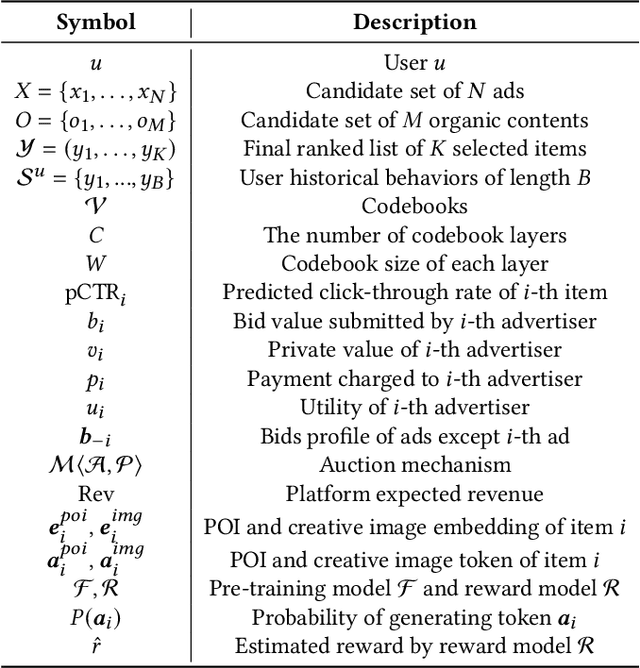
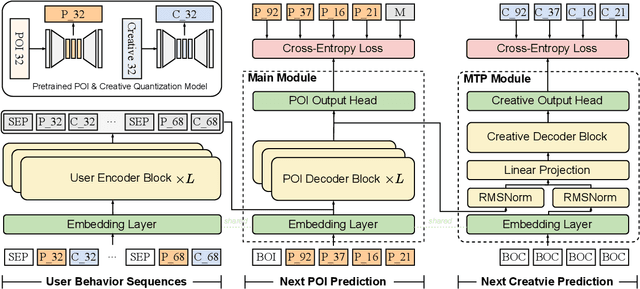
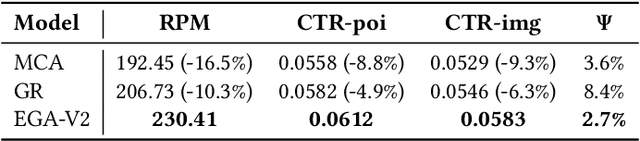
Abstract:Traditional online industrial advertising systems suffer from the limitations of multi-stage cascaded architectures, which often discard high-potential candidates prematurely and distribute decision logic across disconnected modules. While recent generative recommendation approaches provide end-to-end solutions, they fail to address critical advertising requirements of key components for real-world deployment, such as explicit bidding, creative selection, ad allocation, and payment computation. To bridge this gap, we introduce End-to-End Generative Advertising (EGA), the first unified framework that holistically models user interests, point-of-interest (POI) and creative generation, ad allocation, and payment optimization within a single generative model. Our approach employs hierarchical tokenization and multi-token prediction to jointly generate POI recommendations and ad creatives, while a permutation-aware reward model and token-level bidding strategy ensure alignment with both user experiences and advertiser objectives. Additionally, we decouple allocation from payment using a differentiable ex-post regret minimization mechanism, guaranteeing approximate incentive compatibility at the POI level. Through extensive offline evaluations and large-scale online experiments on real-world advertising platforms, we demonstrate that EGA significantly outperforms traditional cascaded systems in both performance and practicality. Our results highlight its potential as a pioneering fully generative advertising solution, paving the way for next-generation industrial ad systems.
One Model to Rank Them All: Unifying Online Advertising with End-to-End Learning
May 26, 2025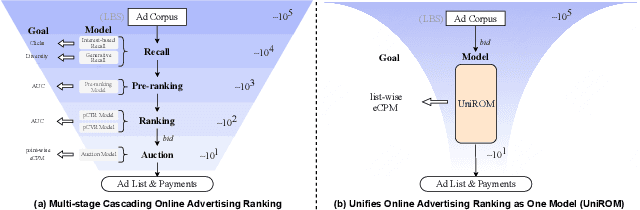
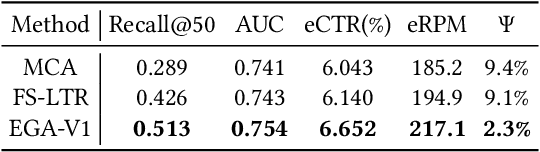
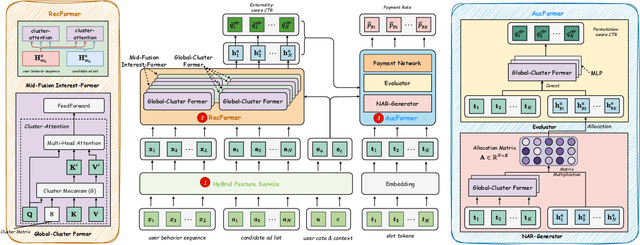
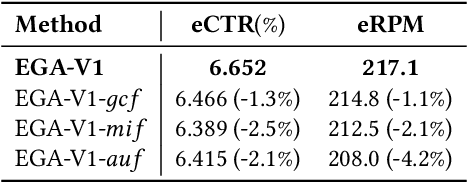
Abstract:Modern industrial advertising systems commonly employ Multi-stage Cascading Architectures (MCA) to balance computational efficiency with ranking accuracy. However, this approach presents two fundamental challenges: (1) performance inconsistencies arising from divergent optimization targets and capability differences between stages, and (2) failure to account for advertisement externalities - the complex interactions between candidate ads during ranking. These limitations ultimately compromise system effectiveness and reduce platform profitability. In this paper, we present UniROM, an end-to-end generative architecture that Unifies online advertising Ranking as One Model. UniROM replaces cascaded stages with a single model to directly generate optimal ad sequences from the full candidate ad corpus in location-based services (LBS). The primary challenges associated with this approach stem from high costs of feature processing and computational bottlenecks in modeling externalities of large-scale candidate pools. To address these challenges, UniROM introduces an algorithm and engine co-designed hybrid feature service to decouple user and ad feature processing, reducing latency while preserving expressiveness. To efficiently extract intra- and cross-sequence mutual information, we propose RecFormer with an innovative cluster-attention mechanism as its core architectural component. Furthermore, we propose a bi-stage training strategy that integrates pre-training with reinforcement learning-based post-training to meet sophisticated platform and advertising objectives. Extensive offline evaluations on public benchmarks and large-scale online A/B testing on industrial advertising platform have demonstrated the superior performance of UniROM over state-of-the-art MCAs.
FHBench: Towards Efficient and Personalized Federated Learning for Multimodal Healthcare
Apr 15, 2025



Abstract:Federated Learning (FL) has emerged as an effective solution for multi-institutional collaborations without sharing patient data, offering a range of methods tailored for diverse applications. However, real-world medical datasets are often multimodal, and computational resources are limited, posing significant challenges for existing FL approaches. Recognizing these limitations, we developed the Federated Healthcare Benchmark(FHBench), a benchmark specifically designed from datasets derived from real-world healthcare applications. FHBench encompasses critical diagnostic tasks across domains such as the nervous, cardiovascular, and respiratory systems and general pathology, providing comprehensive support for multimodal healthcare evaluations and filling a significant gap in existing benchmarks. Building on FHBench, we introduced Efficient Personalized Federated Learning with Adaptive LoRA(EPFL), a personalized FL framework that demonstrates superior efficiency and effectiveness across various healthcare modalities. Our results highlight the robustness of FHBench as a benchmarking tool and the potential of EPFL as an innovative approach to advancing healthcare-focused FL, addressing key limitations of existing methods.
Multi-Class Segmentation of Aortic Branches and Zones in Computed Tomography Angiography: The AortaSeg24 Challenge
Feb 07, 2025



Abstract:Multi-class segmentation of the aorta in computed tomography angiography (CTA) scans is essential for diagnosing and planning complex endovascular treatments for patients with aortic dissections. However, existing methods reduce aortic segmentation to a binary problem, limiting their ability to measure diameters across different branches and zones. Furthermore, no open-source dataset is currently available to support the development of multi-class aortic segmentation methods. To address this gap, we organized the AortaSeg24 MICCAI Challenge, introducing the first dataset of 100 CTA volumes annotated for 23 clinically relevant aortic branches and zones. This dataset was designed to facilitate both model development and validation. The challenge attracted 121 teams worldwide, with participants leveraging state-of-the-art frameworks such as nnU-Net and exploring novel techniques, including cascaded models, data augmentation strategies, and custom loss functions. We evaluated the submitted algorithms using the Dice Similarity Coefficient (DSC) and Normalized Surface Distance (NSD), highlighting the approaches adopted by the top five performing teams. This paper presents the challenge design, dataset details, evaluation metrics, and an in-depth analysis of the top-performing algorithms. The annotated dataset, evaluation code, and implementations of the leading methods are publicly available to support further research. All resources can be accessed at https://aortaseg24.grand-challenge.org.
Outlier Synthesis via Hamiltonian Monte Carlo for Out-of-Distribution Detection
Jan 28, 2025



Abstract:Out-of-distribution (OOD) detection is crucial for developing trustworthy and reliable machine learning systems. Recent advances in training with auxiliary OOD data demonstrate efficacy in enhancing detection capabilities. Nonetheless, these methods heavily rely on acquiring a large pool of high-quality natural outliers. Some prior methods try to alleviate this problem by synthesizing virtual outliers but suffer from either poor quality or high cost due to the monotonous sampling strategy and the heavy-parameterized generative models. In this paper, we overcome all these problems by proposing the Hamiltonian Monte Carlo Outlier Synthesis (HamOS) framework, which views the synthesis process as sampling from Markov chains. Based solely on the in-distribution data, the Markov chains can extensively traverse the feature space and generate diverse and representative outliers, hence exposing the model to miscellaneous potential OOD scenarios. The Hamiltonian Monte Carlo with sampling acceptance rate almost close to 1 also makes our framework enjoy great efficiency. By empirically competing with SOTA baselines on both standard and large-scale benchmarks, we verify the efficacy and efficiency of our proposed HamOS.
Hyperbolic Hypergraph Neural Networks for Multi-Relational Knowledge Hypergraph Representation
Dec 11, 2024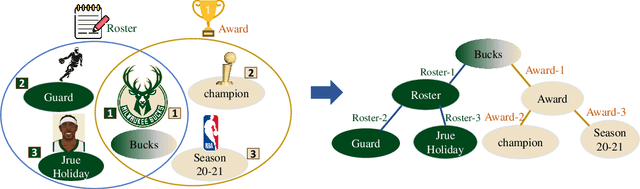
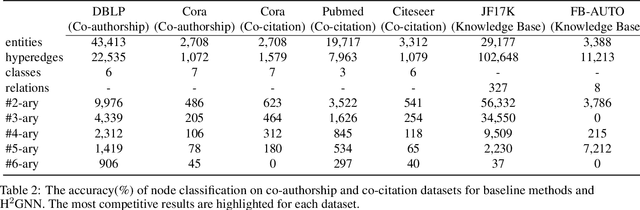
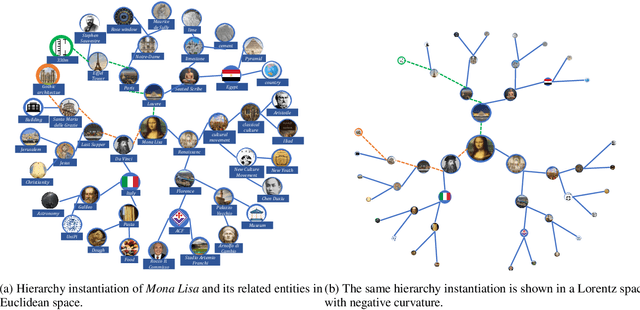
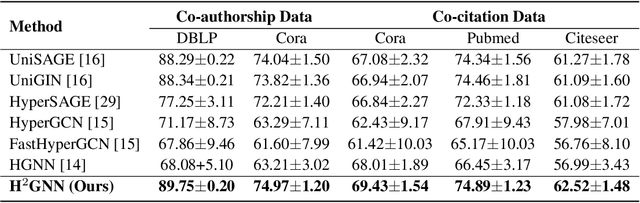
Abstract:Knowledge hypergraphs generalize knowledge graphs using hyperedges to connect multiple entities and depict complicated relations. Existing methods either transform hyperedges into an easier-to-handle set of binary relations or view hyperedges as isolated and ignore their adjacencies. Both approaches have information loss and may potentially lead to the creation of sub-optimal models. To fix these issues, we propose the Hyperbolic Hypergraph Neural Network (H2GNN), whose essential component is the hyper-star message passing, a novel scheme motivated by a lossless expansion of hyperedges into hierarchies. It implements a direct embedding that consciously incorporates adjacent entities, hyper-relations, and entity position-aware information. As the name suggests, H2GNN operates in the hyperbolic space, which is more adept at capturing the tree-like hierarchy. We compare H2GNN with 15 baselines on knowledge hypergraphs, and it outperforms state-of-the-art approaches in both node classification and link prediction tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge