Abul Hasan
Adverse Event Extraction from Discharge Summaries: A New Dataset, Annotation Scheme, and Initial Findings
Jun 17, 2025Abstract:In this work, we present a manually annotated corpus for Adverse Event (AE) extraction from discharge summaries of elderly patients, a population often underrepresented in clinical NLP resources. The dataset includes 14 clinically significant AEs-such as falls, delirium, and intracranial haemorrhage, along with contextual attributes like negation, diagnosis type, and in-hospital occurrence. Uniquely, the annotation schema supports both discontinuous and overlapping entities, addressing challenges rarely tackled in prior work. We evaluate multiple models using FlairNLP across three annotation granularities: fine-grained, coarse-grained, and coarse-grained with negation. While transformer-based models (e.g., BERT-cased) achieve strong performance on document-level coarse-grained extraction (F1 = 0.943), performance drops notably for fine-grained entity-level tasks (e.g., F1 = 0.675), particularly for rare events and complex attributes. These results demonstrate that despite high-level scores, significant challenges remain in detecting underrepresented AEs and capturing nuanced clinical language. Developed within a Trusted Research Environment (TRE), the dataset is available upon request via DataLoch and serves as a robust benchmark for evaluating AE extraction methods and supporting future cross-dataset generalisation.
Harnessing Knowledge Retrieval with Large Language Models for Clinical Report Error Correction
Jun 21, 2024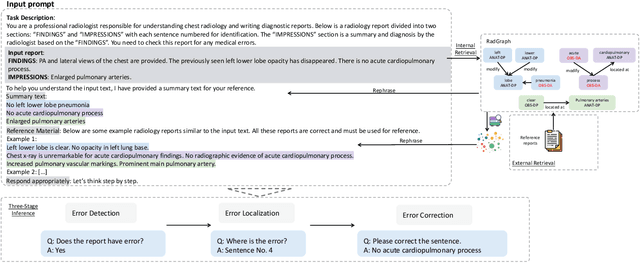



Abstract:This study proposes an approach for error correction in clinical radiology reports, leveraging large language models (LLMs) and retrieval-augmented generation (RAG) techniques. The proposed framework employs internal and external retrieval mechanisms to extract relevant medical entities and relations from the report and external knowledge sources. A three-stage inference process is introduced, decomposing the task into error detection, localization, and correction subtasks, which enhances the explainability and performance of the system. The effectiveness of the approach is evaluated using a benchmark dataset created by corrupting real-world radiology reports with realistic errors, guided by domain experts. Experimental results demonstrate the benefits of the proposed methods, with the combination of internal and external retrieval significantly improving the accuracy of error detection, localization, and correction across various state-of-the-art LLMs. The findings contribute to the development of more robust and reliable error correction systems for clinical documentation.
Infusing clinical knowledge into tokenisers for language models
Jun 20, 2024



Abstract:This study introduces a novel knowledge enhanced tokenisation mechanism, K-Tokeniser, for clinical text processing. Technically, at initialisation stage, K-Tokeniser populates global representations of tokens based on semantic types of domain concepts (such as drugs or diseases) from either a domain ontology like Unified Medical Language System or the training data of the task related corpus. At training or inference stage, sentence level localised context will be utilised for choosing the optimal global token representation to realise the semantic-based tokenisation. To avoid pretraining using the new tokeniser, an embedding initialisation approach is proposed to generate representations for new tokens. Using three transformer-based language models, a comprehensive set of experiments are conducted on four real-world datasets for evaluating K-Tokeniser in a wide range of clinical text analytics tasks including clinical concept and relation extraction, automated clinical coding, clinical phenotype identification, and clinical research article classification. Overall, our models demonstrate consistent improvements over their counterparts in all tasks. In particular, substantial improvements are observed in the automated clinical coding task with 13\% increase on Micro $F_1$ score. Furthermore, K-Tokeniser also shows significant capacities in facilitating quicker converge of language models. Specifically, using K-Tokeniser, the language models would only require 50\% of the training data to achieve the best performance of the baseline tokeniser using all training data in the concept extraction task and less than 20\% of the data for the automated coding task. It is worth mentioning that all these improvements require no pre-training process, making the approach generalisable.
Chain-of-Though (CoT) prompting strategies for medical error detection and correction
Jun 13, 2024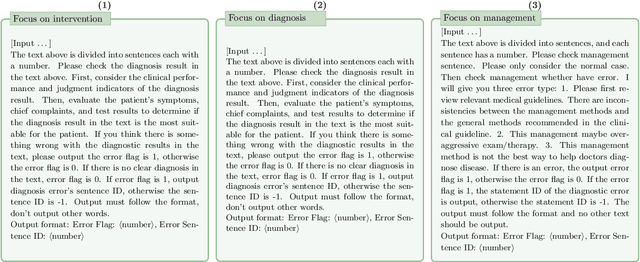
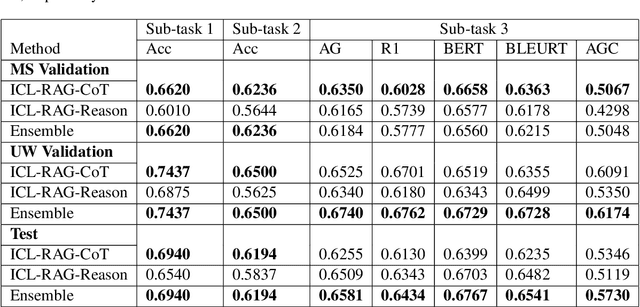

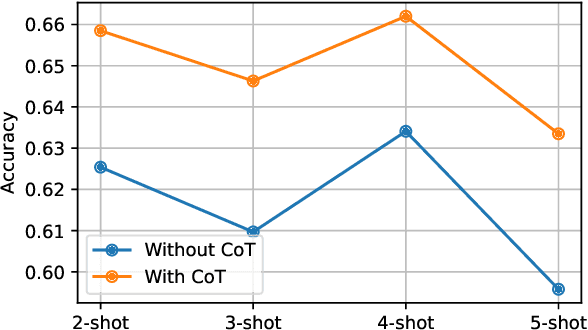
Abstract:This paper describes our submission to the MEDIQA-CORR 2024 shared task for automatically detecting and correcting medical errors in clinical notes. We report results for three methods of few-shot In-Context Learning (ICL) augmented with Chain-of-Thought (CoT) and reason prompts using a large language model (LLM). In the first method, we manually analyse a subset of train and validation dataset to infer three CoT prompts by examining error types in the clinical notes. In the second method, we utilise the training dataset to prompt the LLM to deduce reasons about their correctness or incorrectness. The constructed CoTs and reasons are then augmented with ICL examples to solve the tasks of error detection, span identification, and error correction. Finally, we combine the two methods using a rule-based ensemble method. Across the three sub-tasks, our ensemble method achieves a ranking of 3rd for both sub-task 1 and 2, while securing 7th place in sub-task 3 among all submissions.
RadBARTsum: Domain Specific Adaption of Denoising Sequence-to-Sequence Models for Abstractive Radiology Report Summarization
Jun 05, 2024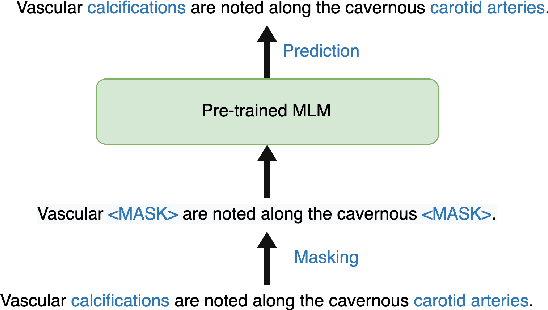
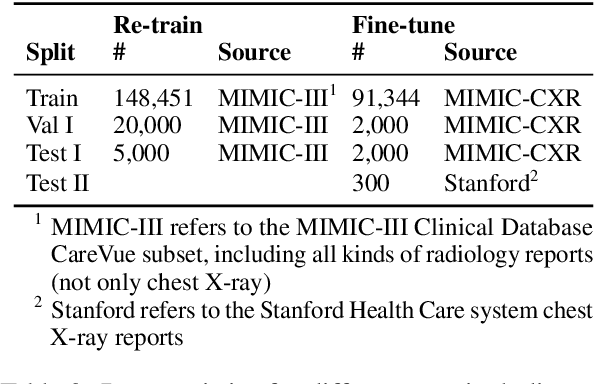
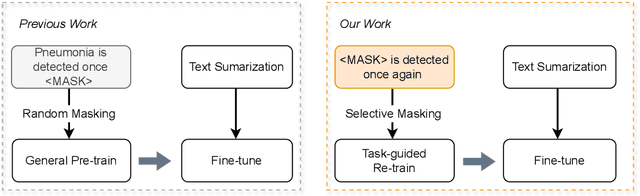
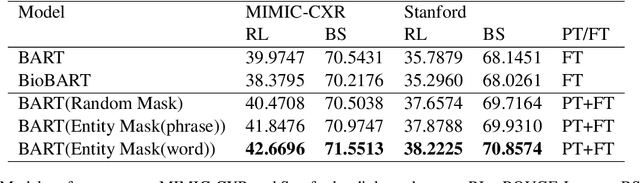
Abstract:Radiology report summarization is a crucial task that can help doctors quickly identify clinically significant findings without the need to review detailed sections of reports. This study proposes RadBARTsum, a domain-specific and ontology facilitated adaptation of the BART model for abstractive radiology report summarization. The approach involves two main steps: 1) re-training the BART model on a large corpus of radiology reports using a novel entity masking strategy to improving biomedical domain knowledge learning, and 2) fine-tuning the model for the summarization task using the Findings and Background sections to predict the Impression section. Experiments are conducted using different masking strategies. Results show that the re-training process with domain knowledge facilitated masking improves performances consistently across various settings. This work contributes a domain-specific generative language model for radiology report summarization and a method for utilising medical knowledge to realise entity masking language model. The proposed approach demonstrates a promising direction of enhancing the efficiency of language models by deepening its understanding of clinical knowledge in radiology reports.
Incorporating Dictionaries into a Neural Network Architecture to Extract COVID-19 Medical Concepts From Social Media
Sep 05, 2023Abstract:We investigate the potential benefit of incorporating dictionary information into a neural network architecture for natural language processing. In particular, we make use of this architecture to extract several concepts related to COVID-19 from an on-line medical forum. We use a sample from the forum to manually curate one dictionary for each concept. In addition, we use MetaMap, which is a tool for extracting biomedical concepts, to identify a small number of semantic concepts. For a supervised concept extraction task on the forum data, our best model achieved a macro $F_1$ score of 90\%. A major difficulty in medical concept extraction is obtaining labelled data from which to build supervised models. We investigate the utility of our models to transfer to data derived from a different source in two ways. First for producing labels via weak learning and second to perform concept extraction. The dataset we use in this case comprises COVID-19 related tweets and we achieve an $F_1$ score 81\% for symptom concept extraction trained on weakly labelled data. The utility of our dictionaries is compared with a COVID-19 symptom dictionary that was constructed directly from Twitter. Further experiments that incorporate BERT and a COVID-19 version of BERTweet demonstrate that the dictionaries provide a commensurate result. Our results show that incorporating small domain dictionaries to deep learning models can improve concept extraction tasks. Moreover, models built using dictionaries generalize well and are transferable to different datasets on a similar task.
Triage and diagnosis of COVID-19 from medical social media
Mar 22, 2021
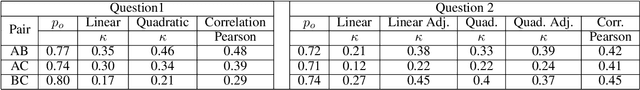
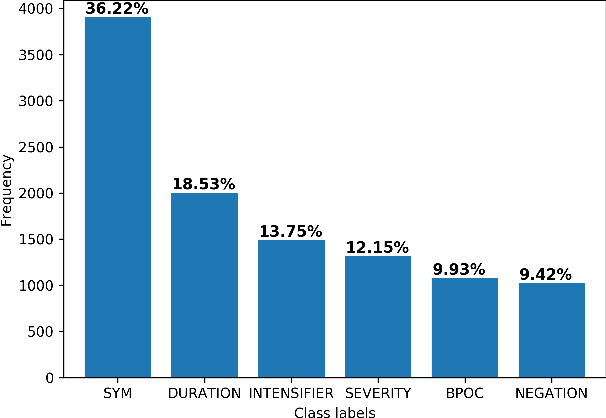
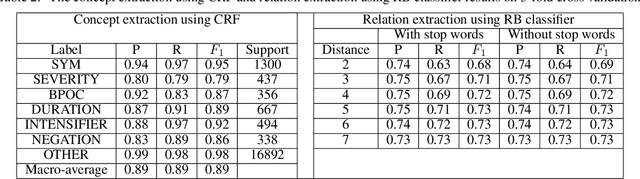
Abstract:Objective: This study aims to develop an end-to-end natural language processing pipeline for triage and diagnosis of COVID-19 from patient-authored social media posts. Materials and Methods: The text processing pipeline first extracts COVID-19 symptoms and related concepts such as severity, duration, negations, and body parts from patients posts using conditional random fields. An unsupervised rule-based algorithm is then applied to establish relations between concepts in the next step of the pipeline. The extracted concepts and relations are subsequently used to construct two different vector representations of each post. These vectors are applied separately to build support vector machine learning models to triage patients into three categories and diagnose them for COVID-19. Results: We report that Macro- and Micro-averaged F_1 scores in the range of 71-96% and 61-87%, respectively, for the triage and diagnosis of COVID-19, when the models are trained on ground truth labelled data. Our experimental results indicate that similar performance can be achieved when the models are trained using predicted labels from concept extraction and rule-based classifiers, thus yielding end-to-end machine learning. Discussion: We highlight important features uncovered by our diagnostic machine learning models and compare them with the most frequent symptoms revealed in another COVID-19 dataset. In particular, we found that the most important features are not always the most frequent ones. Conclusions: Our preliminary results show that it is possible to automatically triage and diagnose patients for COVID-19 from natural language narratives using a machine learning pipeline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge