Honghan Wu
Adverse Event Extraction from Discharge Summaries: A New Dataset, Annotation Scheme, and Initial Findings
Jun 17, 2025Abstract:In this work, we present a manually annotated corpus for Adverse Event (AE) extraction from discharge summaries of elderly patients, a population often underrepresented in clinical NLP resources. The dataset includes 14 clinically significant AEs-such as falls, delirium, and intracranial haemorrhage, along with contextual attributes like negation, diagnosis type, and in-hospital occurrence. Uniquely, the annotation schema supports both discontinuous and overlapping entities, addressing challenges rarely tackled in prior work. We evaluate multiple models using FlairNLP across three annotation granularities: fine-grained, coarse-grained, and coarse-grained with negation. While transformer-based models (e.g., BERT-cased) achieve strong performance on document-level coarse-grained extraction (F1 = 0.943), performance drops notably for fine-grained entity-level tasks (e.g., F1 = 0.675), particularly for rare events and complex attributes. These results demonstrate that despite high-level scores, significant challenges remain in detecting underrepresented AEs and capturing nuanced clinical language. Developed within a Trusted Research Environment (TRE), the dataset is available upon request via DataLoch and serves as a robust benchmark for evaluating AE extraction methods and supporting future cross-dataset generalisation.
Look & Mark: Leveraging Radiologist Eye Fixations and Bounding boxes in Multimodal Large Language Models for Chest X-ray Report Generation
May 28, 2025Abstract:Recent advancements in multimodal Large Language Models (LLMs) have significantly enhanced the automation of medical image analysis, particularly in generating radiology reports from chest X-rays (CXR). However, these models still suffer from hallucinations and clinically significant errors, limiting their reliability in real-world applications. In this study, we propose Look & Mark (L&M), a novel grounding fixation strategy that integrates radiologist eye fixations (Look) and bounding box annotations (Mark) into the LLM prompting framework. Unlike conventional fine-tuning, L&M leverages in-context learning to achieve substantial performance gains without retraining. When evaluated across multiple domain-specific and general-purpose models, L&M demonstrates significant gains, including a 1.2% improvement in overall metrics (A.AVG) for CXR-LLaVA compared to baseline prompting and a remarkable 9.2% boost for LLaVA-Med. General-purpose models also benefit from L&M combined with in-context learning, with LLaVA-OV achieving an 87.3% clinical average performance (C.AVG)-the highest among all models, even surpassing those explicitly trained for CXR report generation. Expert evaluations further confirm that L&M reduces clinically significant errors (by 0.43 average errors per report), such as false predictions and omissions, enhancing both accuracy and reliability. These findings highlight L&M's potential as a scalable and efficient solution for AI-assisted radiology, paving the way for improved diagnostic workflows in low-resource clinical settings.
BioHopR: A Benchmark for Multi-Hop, Multi-Answer Reasoning in Biomedical Domain
May 28, 2025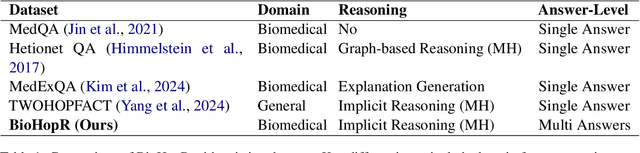
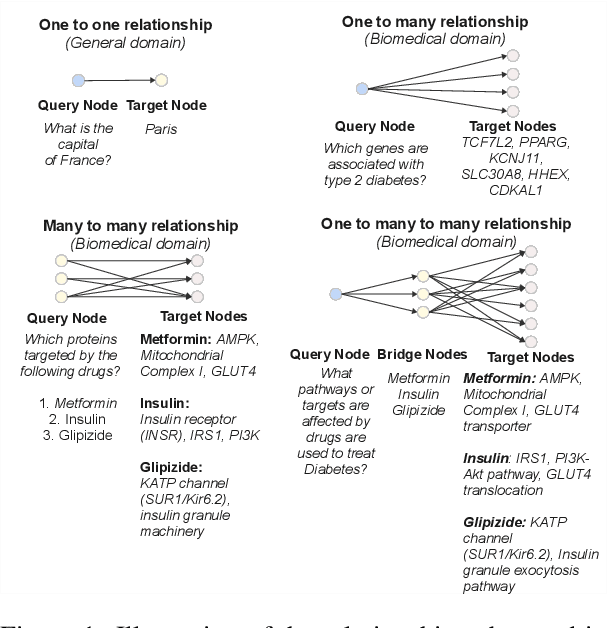
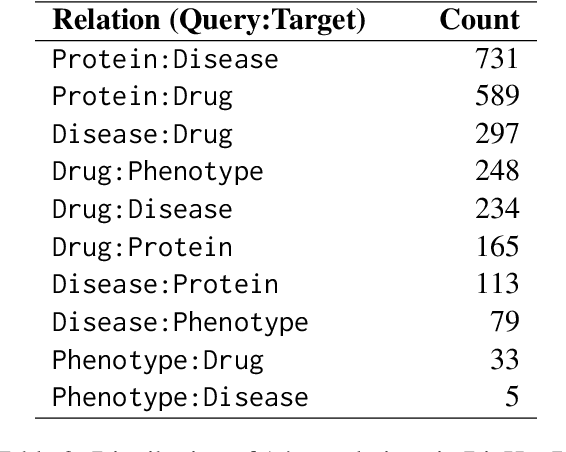
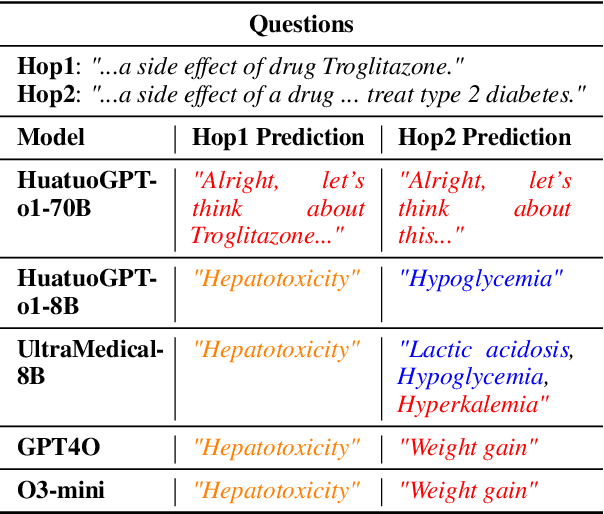
Abstract:Biomedical reasoning often requires traversing interconnected relationships across entities such as drugs, diseases, and proteins. Despite the increasing prominence of large language models (LLMs), existing benchmarks lack the ability to evaluate multi-hop reasoning in the biomedical domain, particularly for queries involving one-to-many and many-to-many relationships. This gap leaves the critical challenges of biomedical multi-hop reasoning underexplored. To address this, we introduce BioHopR, a novel benchmark designed to evaluate multi-hop, multi-answer reasoning in structured biomedical knowledge graphs. Built from the comprehensive PrimeKG, BioHopR includes 1-hop and 2-hop reasoning tasks that reflect real-world biomedical complexities. Evaluations of state-of-the-art models reveal that O3-mini, a proprietary reasoning-focused model, achieves 37.93% precision on 1-hop tasks and 14.57% on 2-hop tasks, outperforming proprietary models such as GPT4O and open-source biomedical models including HuatuoGPT-o1-70B and Llama-3.3-70B. However, all models exhibit significant declines in multi-hop performance, underscoring the challenges of resolving implicit reasoning steps in the biomedical domain. By addressing the lack of benchmarks for multi-hop reasoning in biomedical domain, BioHopR sets a new standard for evaluating reasoning capabilities and highlights critical gaps between proprietary and open-source models while paving the way for future advancements in biomedical LLMs.
Towards deployment-centric multimodal AI beyond vision and language
Apr 04, 2025Abstract:Multimodal artificial intelligence (AI) integrates diverse types of data via machine learning to improve understanding, prediction, and decision-making across disciplines such as healthcare, science, and engineering. However, most multimodal AI advances focus on models for vision and language data, while their deployability remains a key challenge. We advocate a deployment-centric workflow that incorporates deployment constraints early to reduce the likelihood of undeployable solutions, complementing data-centric and model-centric approaches. We also emphasise deeper integration across multiple levels of multimodality and multidisciplinary collaboration to significantly broaden the research scope beyond vision and language. To facilitate this approach, we identify common multimodal-AI-specific challenges shared across disciplines and examine three real-world use cases: pandemic response, self-driving car design, and climate change adaptation, drawing expertise from healthcare, social science, engineering, science, sustainability, and finance. By fostering multidisciplinary dialogue and open research practices, our community can accelerate deployment-centric development for broad societal impact.
IHC-LLMiner: Automated extraction of tumour immunohistochemical profiles from PubMed abstracts using large language models
Apr 01, 2025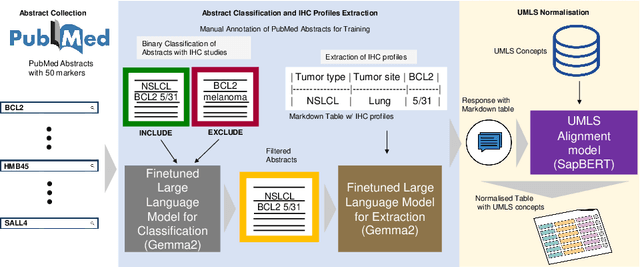
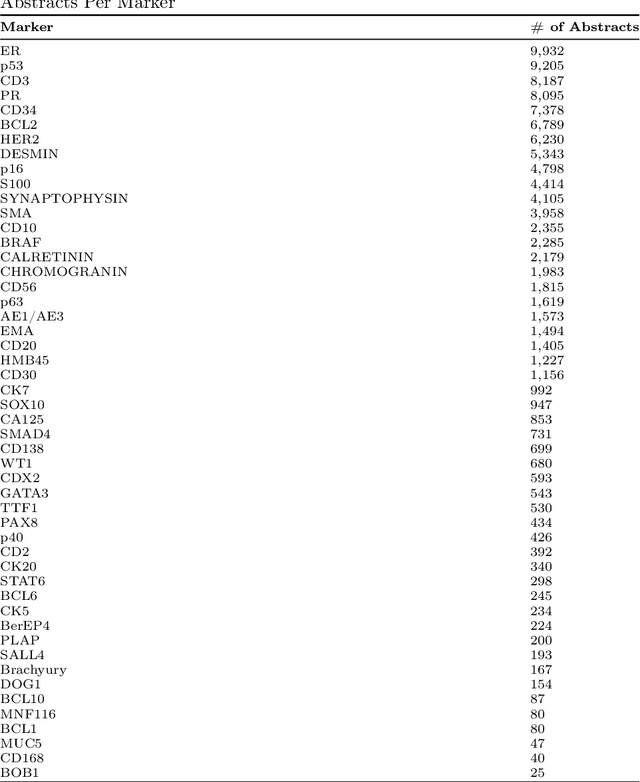
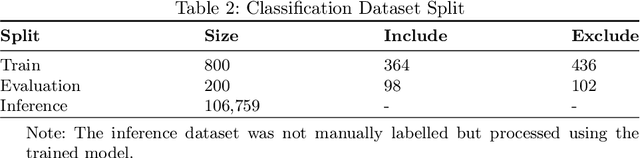
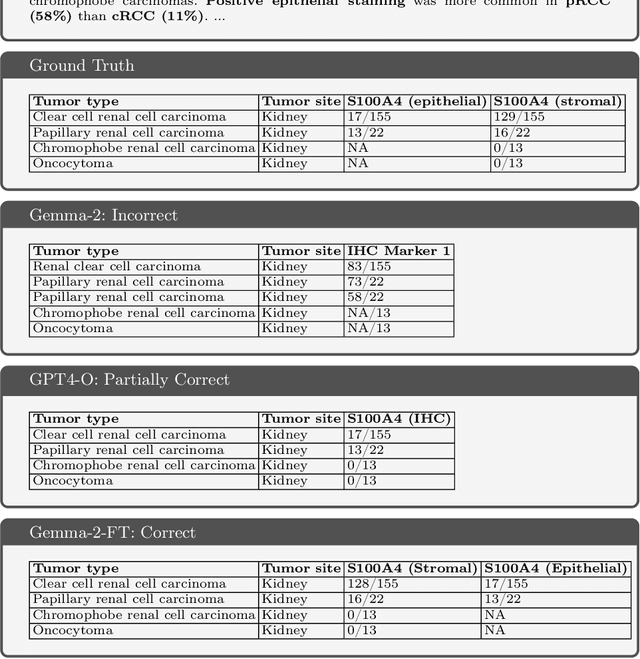
Abstract:Immunohistochemistry (IHC) is essential in diagnostic pathology and biomedical research, offering critical insights into protein expression and tumour biology. This study presents an automated pipeline, IHC-LLMiner, for extracting IHC-tumour profiles from PubMed abstracts, leveraging advanced biomedical text mining. There are two subtasks: abstract classification (include/exclude as relevant) and IHC-tumour profile extraction on relevant included abstracts. The best-performing model, "Gemma-2 finetuned", achieved 91.5% accuracy and an F1 score of 91.4, outperforming GPT4-O by 9.5% accuracy with 5.9 times faster inference time. From an initial dataset of 107,759 abstracts identified for 50 immunohistochemical markers, the classification task identified 30,481 relevant abstracts (Include) using the Gemma-2 finetuned model. For IHC-tumour profile extraction, the Gemma-2 finetuned model achieved the best performance with 63.3% Correct outputs. Extracted IHC-tumour profiles (tumour types and markers) were normalised to Unified Medical Language System (UMLS) concepts to ensure consistency and facilitate IHC-tumour profile landscape analysis. The extracted IHC-tumour profiles demonstrated excellent concordance with available online summary data and provided considerable added value in terms of both missing IHC-tumour profiles and quantitative assessments. Our proposed LLM based pipeline provides a practical solution for large-scale IHC-tumour profile data mining, enhancing the accessibility and utility of such data for research and clinical applications as well as enabling the generation of quantitative and structured data to support cancer-specific knowledge base development. Models and training datasets are available at https://github.com/knowlab/IHC-LLMiner.
RiskAgent: Autonomous Medical AI Copilot for Generalist Risk Prediction
Mar 05, 2025Abstract:The application of Large Language Models (LLMs) to various clinical applications has attracted growing research attention. However, real-world clinical decision-making differs significantly from the standardized, exam-style scenarios commonly used in current efforts. In this paper, we present the RiskAgent system to perform a broad range of medical risk predictions, covering over 387 risk scenarios across diverse complex diseases, e.g., cardiovascular disease and cancer. RiskAgent is designed to collaborate with hundreds of clinical decision tools, i.e., risk calculators and scoring systems that are supported by evidence-based medicine. To evaluate our method, we have built the first benchmark MedRisk specialized for risk prediction, including 12,352 questions spanning 154 diseases, 86 symptoms, 50 specialties, and 24 organ systems. The results show that our RiskAgent, with 8 billion model parameters, achieves 76.33% accuracy, outperforming the most recent commercial LLMs, o1, o3-mini, and GPT-4.5, and doubling the 38.39% accuracy of GPT-4o. On rare diseases, e.g., Idiopathic Pulmonary Fibrosis (IPF), RiskAgent outperforms o1 and GPT-4.5 by 27.27% and 45.46% accuracy, respectively. Finally, we further conduct a generalization evaluation on an external evidence-based diagnosis benchmark and show that our RiskAgent achieves the best results. These encouraging results demonstrate the great potential of our solution for diverse diagnosis domains. To improve the adaptability of our model in different scenarios, we have built and open-sourced a family of models ranging from 1 billion to 70 billion parameters. Our code, data, and models are all available at https://github.com/AI-in-Health/RiskAgent.
Harnessing Knowledge Retrieval with Large Language Models for Clinical Report Error Correction
Jun 21, 2024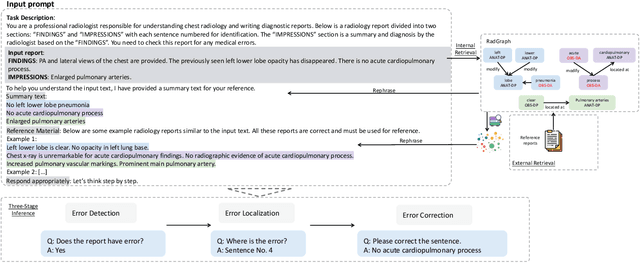



Abstract:This study proposes an approach for error correction in clinical radiology reports, leveraging large language models (LLMs) and retrieval-augmented generation (RAG) techniques. The proposed framework employs internal and external retrieval mechanisms to extract relevant medical entities and relations from the report and external knowledge sources. A three-stage inference process is introduced, decomposing the task into error detection, localization, and correction subtasks, which enhances the explainability and performance of the system. The effectiveness of the approach is evaluated using a benchmark dataset created by corrupting real-world radiology reports with realistic errors, guided by domain experts. Experimental results demonstrate the benefits of the proposed methods, with the combination of internal and external retrieval significantly improving the accuracy of error detection, localization, and correction across various state-of-the-art LLMs. The findings contribute to the development of more robust and reliable error correction systems for clinical documentation.
Infusing clinical knowledge into tokenisers for language models
Jun 20, 2024



Abstract:This study introduces a novel knowledge enhanced tokenisation mechanism, K-Tokeniser, for clinical text processing. Technically, at initialisation stage, K-Tokeniser populates global representations of tokens based on semantic types of domain concepts (such as drugs or diseases) from either a domain ontology like Unified Medical Language System or the training data of the task related corpus. At training or inference stage, sentence level localised context will be utilised for choosing the optimal global token representation to realise the semantic-based tokenisation. To avoid pretraining using the new tokeniser, an embedding initialisation approach is proposed to generate representations for new tokens. Using three transformer-based language models, a comprehensive set of experiments are conducted on four real-world datasets for evaluating K-Tokeniser in a wide range of clinical text analytics tasks including clinical concept and relation extraction, automated clinical coding, clinical phenotype identification, and clinical research article classification. Overall, our models demonstrate consistent improvements over their counterparts in all tasks. In particular, substantial improvements are observed in the automated clinical coding task with 13\% increase on Micro $F_1$ score. Furthermore, K-Tokeniser also shows significant capacities in facilitating quicker converge of language models. Specifically, using K-Tokeniser, the language models would only require 50\% of the training data to achieve the best performance of the baseline tokeniser using all training data in the concept extraction task and less than 20\% of the data for the automated coding task. It is worth mentioning that all these improvements require no pre-training process, making the approach generalisable.
Chain-of-Though (CoT) prompting strategies for medical error detection and correction
Jun 13, 2024
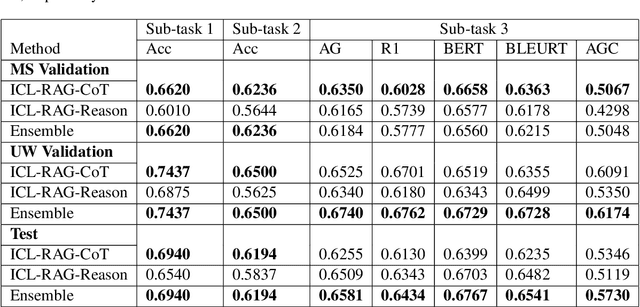

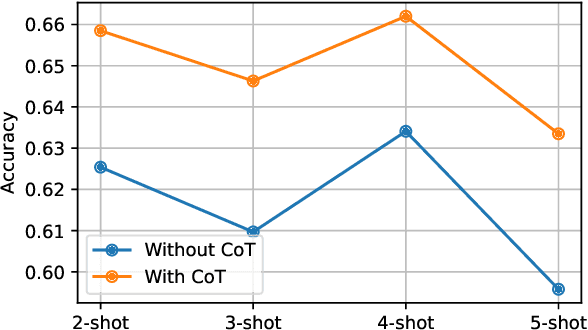
Abstract:This paper describes our submission to the MEDIQA-CORR 2024 shared task for automatically detecting and correcting medical errors in clinical notes. We report results for three methods of few-shot In-Context Learning (ICL) augmented with Chain-of-Thought (CoT) and reason prompts using a large language model (LLM). In the first method, we manually analyse a subset of train and validation dataset to infer three CoT prompts by examining error types in the clinical notes. In the second method, we utilise the training dataset to prompt the LLM to deduce reasons about their correctness or incorrectness. The constructed CoTs and reasons are then augmented with ICL examples to solve the tasks of error detection, span identification, and error correction. Finally, we combine the two methods using a rule-based ensemble method. Across the three sub-tasks, our ensemble method achieves a ranking of 3rd for both sub-task 1 and 2, while securing 7th place in sub-task 3 among all submissions.
MedExQA: Medical Question Answering Benchmark with Multiple Explanations
Jun 10, 2024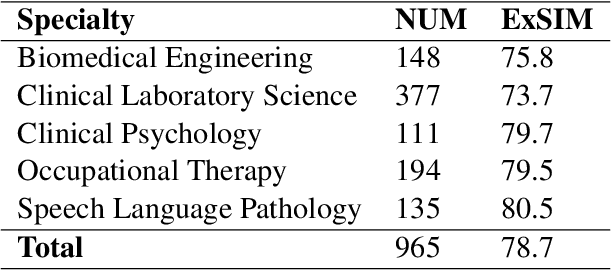
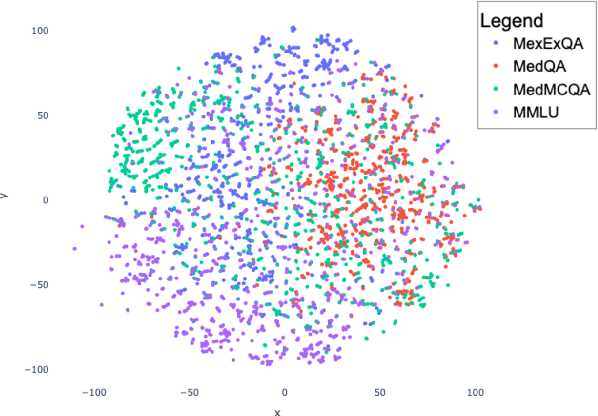
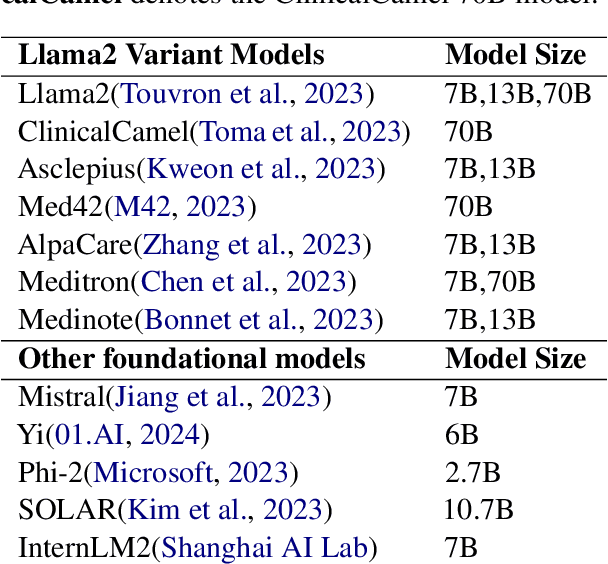
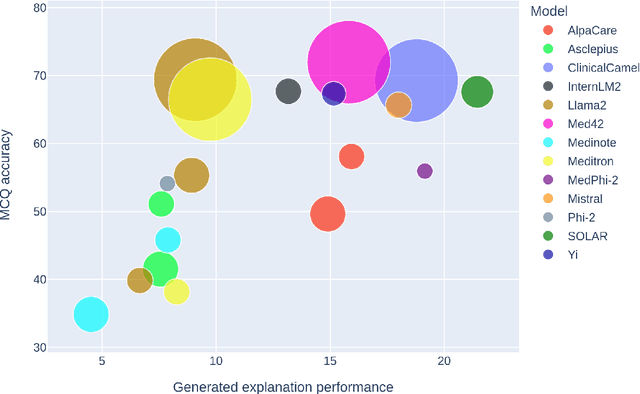
Abstract:This paper introduces MedExQA, a novel benchmark in medical question-answering, to evaluate large language models' (LLMs) understanding of medical knowledge through explanations. By constructing datasets across five distinct medical specialties that are underrepresented in current datasets and further incorporating multiple explanations for each question-answer pair, we address a major gap in current medical QA benchmarks which is the absence of comprehensive assessments of LLMs' ability to generate nuanced medical explanations. Our work highlights the importance of explainability in medical LLMs, proposes an effective methodology for evaluating models beyond classification accuracy, and sheds light on one specific domain, speech language pathology, where current LLMs including GPT4 lack good understanding. Our results show generation evaluation with multiple explanations aligns better with human assessment, highlighting an opportunity for a more robust automated comprehension assessment for LLMs. To diversify open-source medical LLMs (currently mostly based on Llama2), this work also proposes a new medical model, MedPhi-2, based on Phi-2 (2.7B). The model outperformed medical LLMs based on Llama2-70B in generating explanations, showing its effectiveness in the resource-constrained medical domain. We will share our benchmark datasets and the trained model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge