Xianyuan Liu
Towards deployment-centric multimodal AI beyond vision and language
Apr 04, 2025Abstract:Multimodal artificial intelligence (AI) integrates diverse types of data via machine learning to improve understanding, prediction, and decision-making across disciplines such as healthcare, science, and engineering. However, most multimodal AI advances focus on models for vision and language data, while their deployability remains a key challenge. We advocate a deployment-centric workflow that incorporates deployment constraints early to reduce the likelihood of undeployable solutions, complementing data-centric and model-centric approaches. We also emphasise deeper integration across multiple levels of multimodality and multidisciplinary collaboration to significantly broaden the research scope beyond vision and language. To facilitate this approach, we identify common multimodal-AI-specific challenges shared across disciplines and examine three real-world use cases: pandemic response, self-driving car design, and climate change adaptation, drawing expertise from healthcare, social science, engineering, science, sustainability, and finance. By fostering multidisciplinary dialogue and open research practices, our community can accelerate deployment-centric development for broad societal impact.
Interpretable Multimodal Learning for Tumor Protein-Metal Binding: Progress, Challenges, and Perspectives
Apr 04, 2025Abstract:In cancer therapeutics, protein-metal binding mechanisms critically govern drug pharmacokinetics and targeting efficacy, thereby fundamentally shaping the rational design of anticancer metallodrugs. While conventional laboratory methods used to study such mechanisms are often costly, low throughput, and limited in capturing dynamic biological processes, machine learning (ML) has emerged as a promising alternative. Despite increasing efforts to develop protein-metal binding datasets and ML algorithms, the application of ML in tumor protein-metal binding remains limited. Key challenges include a shortage of high-quality, tumor-specific datasets, insufficient consideration of multiple data modalities, and the complexity of interpreting results due to the ''black box'' nature of complex ML models. This paper summarizes recent progress and ongoing challenges in using ML to predict tumor protein-metal binding, focusing on data, modeling, and interpretability. We present multimodal protein-metal binding datasets and outline strategies for acquiring, curating, and preprocessing them for training ML models. Moreover, we explore the complementary value provided by different data modalities and examine methods for their integration. We also review approaches for improving model interpretability to support more trustworthy decisions in cancer research. Finally, we offer our perspective on research opportunities and propose strategies to address the scarcity of tumor protein data and the limited number of predictive models for tumor protein-metal binding. We also highlight two promising directions for effective metal-based drug design: integrating protein-protein interaction data to provide structural insights into metal-binding events and predicting structural changes in tumor proteins after metal binding.
Co-evolution-based Metal-binding Residue Prediction with Graph Neural Networks
Feb 22, 2025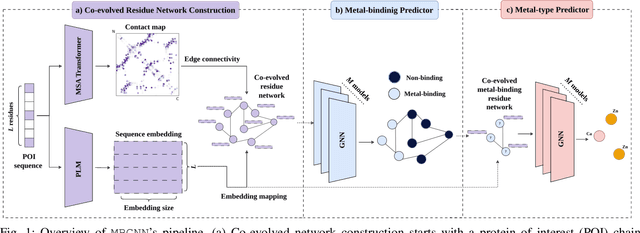
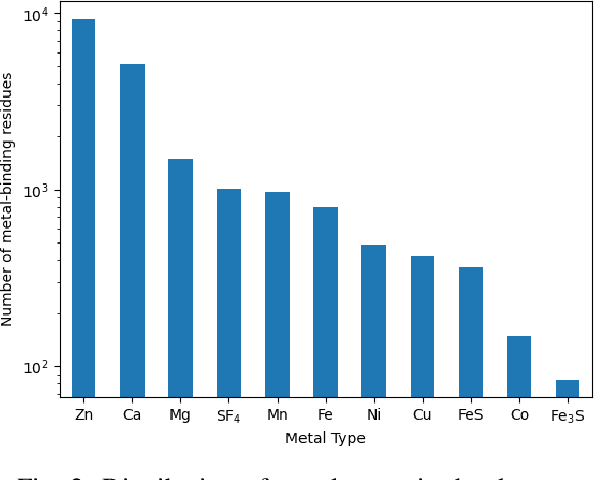
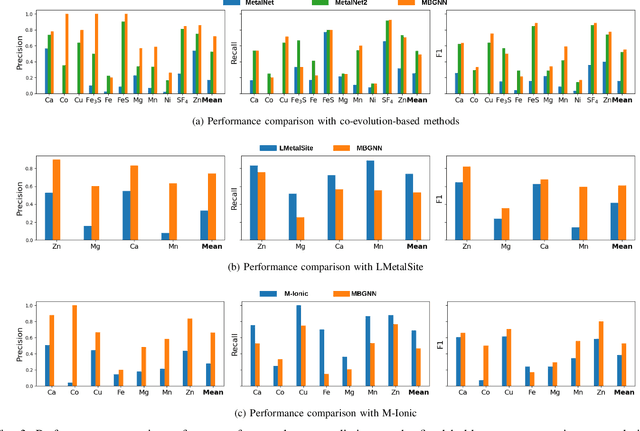
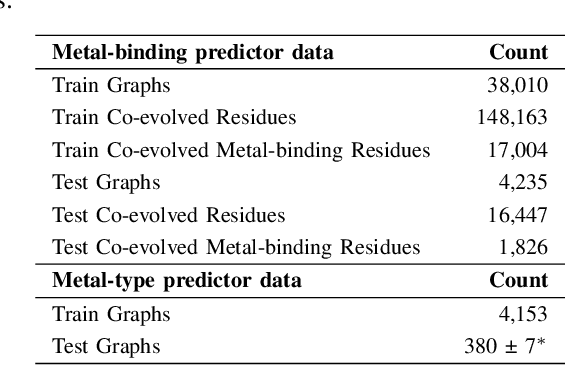
Abstract:In computational structural biology, predicting metal-binding sites and their corresponding metal types is challenging due to the complexity of protein structures and interactions. Conventional sequence- and structure-based prediction approaches cannot capture the complex evolutionary relationships driving these interactions to facilitate understanding, while recent co-evolution-based approaches do not fully consider the entire structure of the co-evolved residue network. In this paper, we introduce MBGNN (Metal-Binding Graph Neural Network) that utilizes the entire co-evolved residue network and effectively captures the complex dependencies within protein structures via graph neural networks to enhance the prediction of co-evolved metal-binding residues and their associated metal types. Experimental results on a public dataset show that MBGNN outperforms existing co-evolution-based metal-binding prediction methods, and it is also competitive against recent sequence-based methods, showing the potential of integrating co-evolutionary insights with advanced machine learning to deepen our understanding of protein-metal interactions. The MBGNN code is publicly available at https://github.com/SRastegari/MBGNN.
Classifying the Stoichiometry of Virus-like Particles with Interpretable Machine Learning
Feb 17, 2025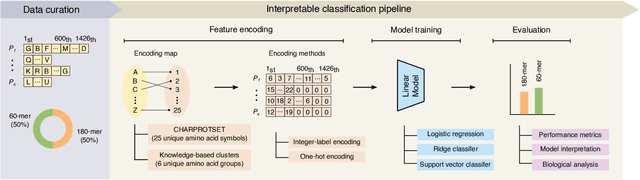
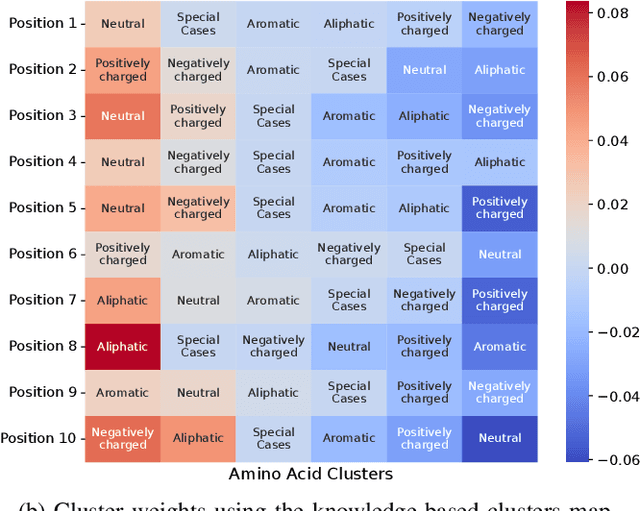
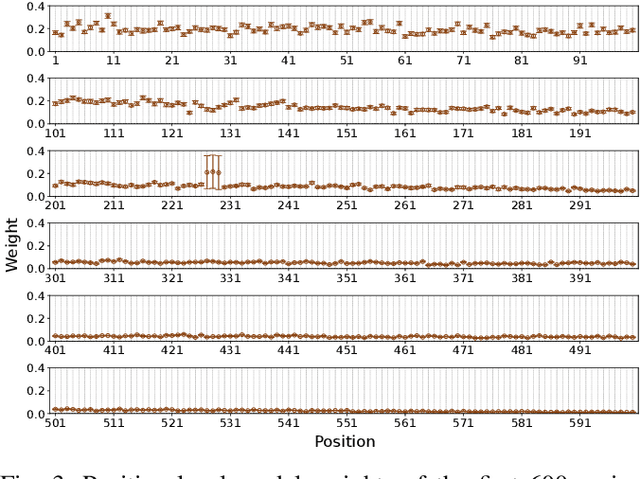
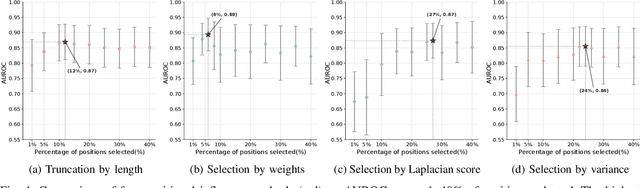
Abstract:Virus-like particles (VLPs) are valuable for vaccine development due to their immune-triggering properties. Understanding their stoichiometry, the number of protein subunits to form a VLP, is critical for vaccine optimisation. However, current experimental methods to determine stoichiometry are time-consuming and require highly purified proteins. To efficiently classify stoichiometry classes in proteins, we curate a new dataset and propose an interpretable, data-driven pipeline leveraging linear machine learning models. We also explore the impact of feature encoding on model performance and interpretability, as well as methods to identify key protein sequence features influencing classification. The evaluation of our pipeline demonstrates that it can classify stoichiometry while revealing protein features that possibly influence VLP assembly. The data and code used in this work are publicly available at https://github.com/Shef-AIRE/StoicIML.
Mask prior-guided denoising diffusion improves inverse protein folding
Dec 10, 2024



Abstract:Inverse protein folding generates valid amino acid sequences that can fold into a desired protein structure, with recent deep-learning advances showing significant potential and competitive performance. However, challenges remain in predicting highly uncertain regions, such as those with loops and disorders. To tackle such low-confidence residue prediction, we propose a \textbf{Ma}sk \textbf{p}rior-guided denoising \textbf{Diff}usion (\textbf{MapDiff}) framework that accurately captures both structural and residue interactions for inverse protein folding. MapDiff is a discrete diffusion probabilistic model that iteratively generates amino acid sequences with reduced noise, conditioned on a given protein backbone. To incorporate structural and residue interactions, we develop a graph-based denoising network with a mask prior pre-training strategy. Moreover, in the generative process, we combine the denoising diffusion implicit model with Monte-Carlo dropout to improve uncertainty estimation. Evaluation on four challenging sequence design benchmarks shows that MapDiff significantly outperforms state-of-the-art methods. Furthermore, the in-silico sequences generated by MapDiff closely resemble the physico-chemical and structural characteristics of native proteins across different protein families and architectures.
DVPE: Divided View Position Embedding for Multi-View 3D Object Detection
Jul 24, 2024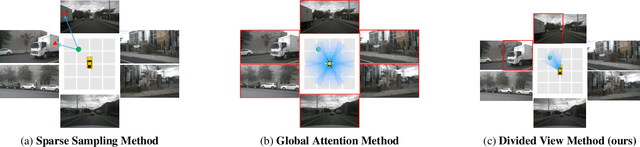
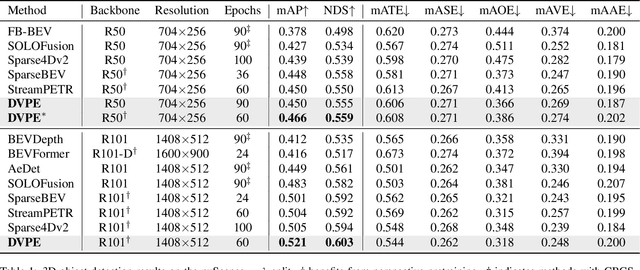
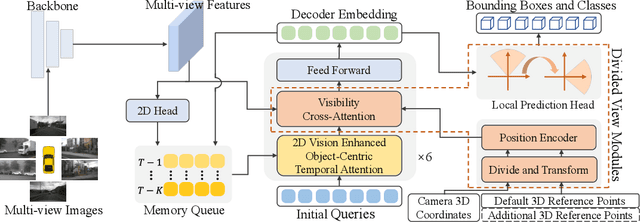
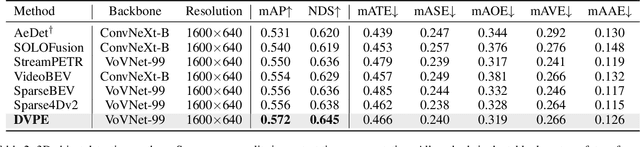
Abstract:Sparse query-based paradigms have achieved significant success in multi-view 3D detection for autonomous vehicles. Current research faces challenges in balancing between enlarging receptive fields and reducing interference when aggregating multi-view features. Moreover, different poses of cameras present challenges in training global attention models. To address these problems, this paper proposes a divided view method, in which features are modeled globally via the visibility crossattention mechanism, but interact only with partial features in a divided local virtual space. This effectively reduces interference from other irrelevant features and alleviates the training difficulties of the transformer by decoupling the position embedding from camera poses. Additionally, 2D historical RoI features are incorporated into the object-centric temporal modeling to utilize highlevel visual semantic information. The model is trained using a one-to-many assignment strategy to facilitate stability. Our framework, named DVPE, achieves state-of-the-art performance (57.2% mAP and 64.5% NDS) on the nuScenes test set. Codes will be available at https://github.com/dop0/DVPE.
Multimodal Variational Autoencoder for Low-cost Cardiac Hemodynamics Instability Detection
Mar 20, 2024
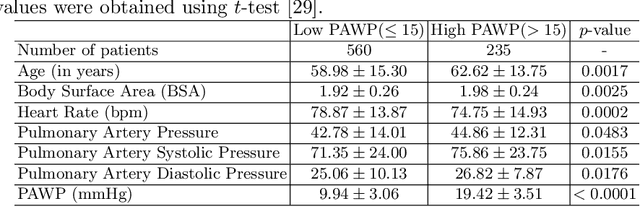
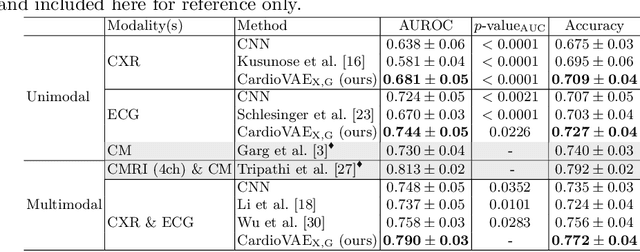
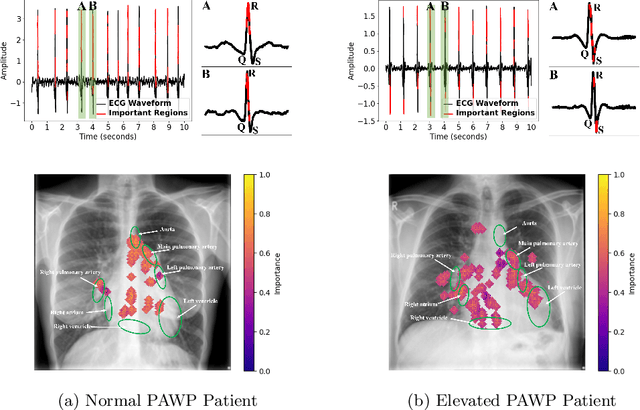
Abstract:Recent advancements in non-invasive detection of cardiac hemodynamic instability (CHDI) primarily focus on applying machine learning techniques to a single data modality, e.g. cardiac magnetic resonance imaging (MRI). Despite their potential, these approaches often fall short especially when the size of labeled patient data is limited, a common challenge in the medical domain. Furthermore, only a few studies have explored multimodal methods to study CHDI, which mostly rely on costly modalities such as cardiac MRI and echocardiogram. In response to these limitations, we propose a novel multimodal variational autoencoder ($\text{CardioVAE}_\text{X,G}$) to integrate low-cost chest X-ray (CXR) and electrocardiogram (ECG) modalities with pre-training on a large unlabeled dataset. Specifically, $\text{CardioVAE}_\text{X,G}$ introduces a novel tri-stream pre-training strategy to learn both shared and modality-specific features, thus enabling fine-tuning with both unimodal and multimodal datasets. We pre-train $\text{CardioVAE}_\text{X,G}$ on a large, unlabeled dataset of $50,982$ subjects from a subset of MIMIC database and then fine-tune the pre-trained model on a labeled dataset of $795$ subjects from the ASPIRE registry. Comprehensive evaluations against existing methods show that $\text{CardioVAE}_\text{X,G}$ offers promising performance (AUROC $=0.79$ and Accuracy $=0.77$), representing a significant step forward in non-invasive prediction of CHDI. Our model also excels in producing fine interpretations of predictions directly associated with clinical features, thereby supporting clinical decision-making.
MeDSLIP: Medical Dual-Stream Language-Image Pre-training for Fine-grained Alignment
Mar 15, 2024Abstract:Vision-language pre-training (VLP) models have shown significant advancements in the medical domain. Yet, most VLP models align raw reports to images at a very coarse level, without modeling fine-grained relationships between anatomical and pathological concepts outlined in reports and the corresponding semantic counterparts in images. To address this problem, we propose a Medical Dual-Stream Language-Image Pre-training (MeDSLIP) framework. Specifically, MeDSLIP establishes vision-language fine-grained alignments via disentangling visual and textual representations into anatomy-relevant and pathology-relevant streams. Moreover, a novel vision-language Prototypical Contr-astive Learning (ProtoCL) method is adopted in MeDSLIP to enhance the alignment within the anatomical and pathological streams. MeDSLIP further employs cross-stream Intra-image Contrastive Learning (ICL) to ensure the consistent coexistence of paired anatomical and pathological concepts within the same image. Such a cross-stream regularization encourages the model to exploit the synchrony between two streams for a more comprehensive representation learning. MeDSLIP is evaluated under zero-shot and supervised fine-tuning settings on three public datasets: NIH CXR14, RSNA Pneumonia, and SIIM-ACR Pneumothorax. Under these settings, MeDSLIP outperforms six leading CNN-based models on classification, grounding, and segmentation tasks.
Geometry-aware Line Graph Transformer Pre-training for Molecular Property Prediction
Sep 01, 2023



Abstract:Molecular property prediction with deep learning has gained much attention over the past years. Owing to the scarcity of labeled molecules, there has been growing interest in self-supervised learning methods that learn generalizable molecular representations from unlabeled data. Molecules are typically treated as 2D topological graphs in modeling, but it has been discovered that their 3D geometry is of great importance in determining molecular functionalities. In this paper, we propose the Geometry-aware line graph transformer (Galformer) pre-training, a novel self-supervised learning framework that aims to enhance molecular representation learning with 2D and 3D modalities. Specifically, we first design a dual-modality line graph transformer backbone to encode the topological and geometric information of a molecule. The designed backbone incorporates effective structural encodings to capture graph structures from both modalities. Then we devise two complementary pre-training tasks at the inter and intra-modality levels. These tasks provide properly supervised information and extract discriminative 2D and 3D knowledge from unlabeled molecules. Finally, we evaluate Galformer against six state-of-the-art baselines on twelve property prediction benchmarks via downstream fine-tuning. Experimental results show that Galformer consistently outperforms all baselines on both classification and regression tasks, demonstrating its effectiveness.
Channel-Temporal Attention for First-Person Video Domain Adaptation
Aug 19, 2021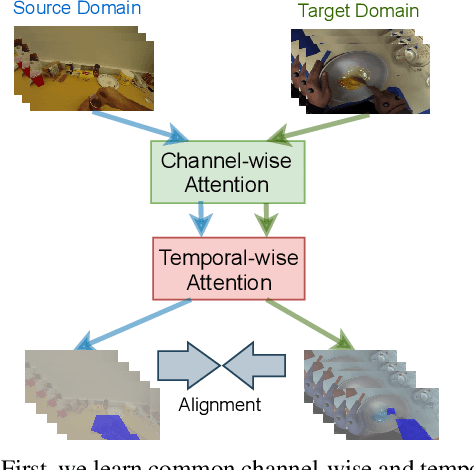
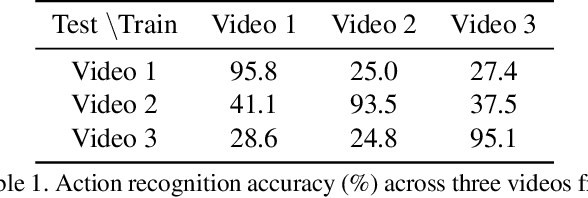
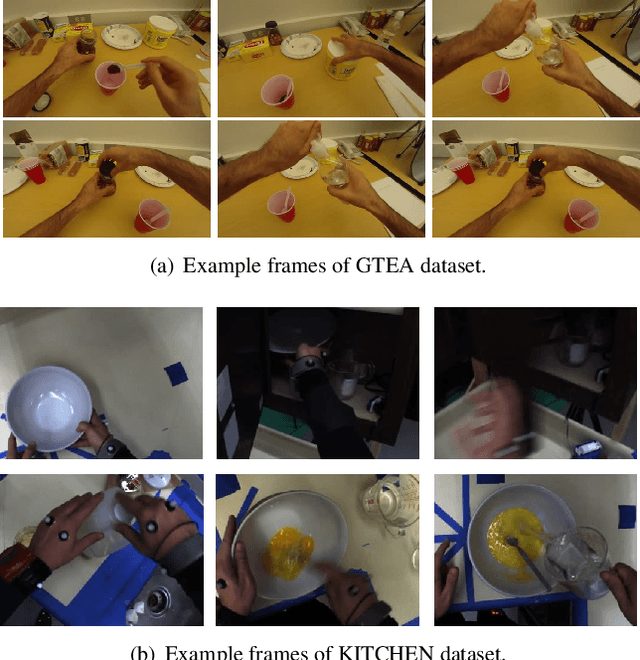
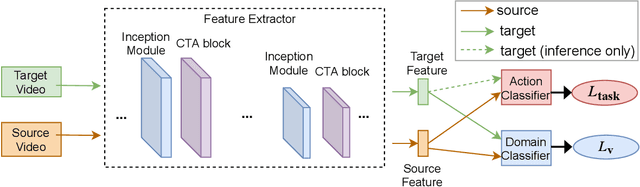
Abstract:Unsupervised Domain Adaptation (UDA) can transfer knowledge from labeled source data to unlabeled target data of the same categories. However, UDA for first-person action recognition is an under-explored problem, with lack of datasets and limited consideration of first-person video characteristics. This paper focuses on addressing this problem. Firstly, we propose two small-scale first-person video domain adaptation datasets: ADL$_{small}$ and GTEA-KITCHEN. Secondly, we introduce channel-temporal attention blocks to capture the channel-wise and temporal-wise relationships and model their inter-dependencies important to first-person vision. Finally, we propose a Channel-Temporal Attention Network (CTAN) to integrate these blocks into existing architectures. CTAN outperforms baselines on the two proposed datasets and one existing dataset EPIC$_{cvpr20}$.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge